|
|
|
|
|
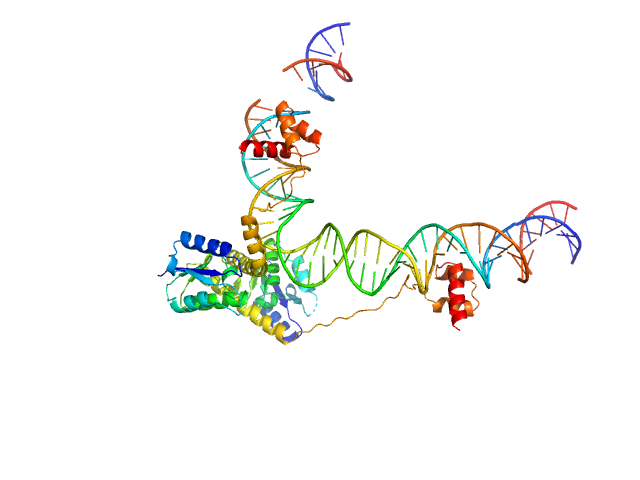
| Sample: |
Transposon Tn3 resolvase dimer, 43 kDa Escherichia coli protein
Tn3 res, resolvase binding site II monomer, 32 kDa DNA
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 10 mM MgCl2, 1% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 6
|
Structural basis for topological regulation of Tn3 resolvase.
Nucleic Acids Res (2022)
Montaño SP, Rowland SJ, Fuller JR, Burke ME, MacDonald AI, Boocock MR, Stark WM, Rice PA
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 4
|
New insights into P2X7 receptor regulation: Ca2+-calmodulin and GDP bind to the soluble P2X7 ballast domain.
J Biol Chem :102495 (2022)
Sander S, Müller I, Alai MG, Nicke A, Tidow H
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
P2X purinoceptor 7 trimer, 74 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 4
|
New insights into P2X7 receptor regulation: Ca2+-calmodulin and GDP bind to the soluble P2X7 ballast domain.
J Biol Chem :102495 (2022)
Sander S, Müller I, Alai MG, Nicke A, Tidow H
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
P2X purinoceptor 7 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 4
|
New insights into P2X7 receptor regulation: Ca2+-calmodulin and GDP bind to the soluble P2X7 ballast domain.
J Biol Chem :102495 (2022)
Sander S, Müller I, Alai MG, Nicke A, Tidow H
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
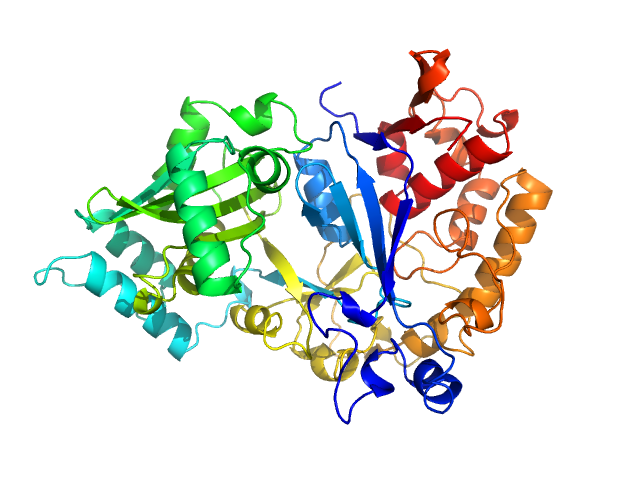
| Sample: |
Arginyl-tRNA--protein transferase 1 monomer, 60 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
2% glycerol, 100 mM KCl, 50 mM Tris-HCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 12
|
The Structure of Saccharomyces cerevisiae Arginyltransferase 1 (ATE1).
J Mol Biol 434(21):167816 (2022)
Van V, Ejimogu NE, Bui TS, Smith AT
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Testis-expressed protein 12 dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 10
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Testis-expressed protein 12 tetramer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 10
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Testis-expressed protein 12 (L110E, F114E, I117E, L121E) tetramer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 15
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Testis-expressed protein 12 (F102A, F109E, V116A) tetramer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 27
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Testis-expressed protein 12 (F109E) dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 13
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
30 |
nm3 |
|
|