|
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) dimer, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 250 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) dimer, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 500 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) dimer, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1000 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) tetramer, 88 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 250 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
6.3 |
nm |
| Dmax |
24.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) tetramer, 88 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 500 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1a (Non-structural protein 8, SARS-CoV-2) tetramer, 88 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1000 mM NaCl, pH: 7.4 |
| Experiment: |
SANS
data collected at BL01-Small Angle Neutron Scattering, China Spallation Neutron Source on 2021 Jul 19
|
Multiscale characterization reveals oligomerization dependent phase separation of primer-independent RNA polymerase nsp8 from SARS-CoV-2.
Commun Biol 5(1):925 (2022)
Xu J, Jiang X, Zhang Y, Dong Y, Ma C, Jiang H, Zuo T, Chen R, Ke Y, Cheng H, Wang H, Liu J
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
|
|
|
|
|
|
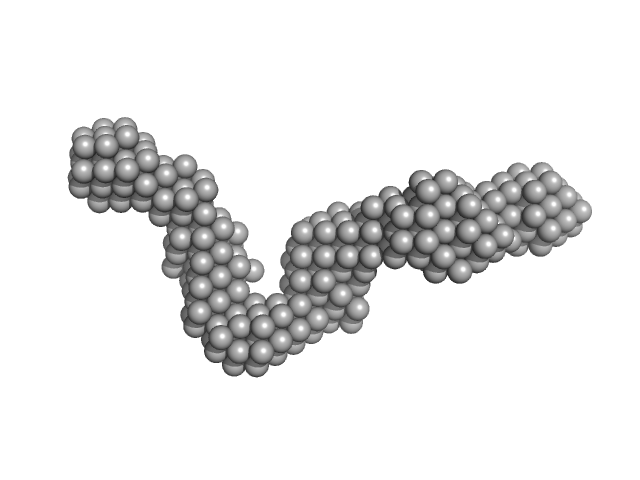
|
| Sample: |
40-mer single stranded inhibitory DNA monomer, 12 kDa DNA
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 8
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA dC->dU-editing enzyme APOBEC-3G tetramer, 186 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA dC->dU-editing enzyme APOBEC-3G tetramer, 186 kDa Homo sapiens protein
40-mer single stranded inhibitory DNA dimer, 24 kDa DNA
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
395 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
40-mer single stranded inhibitory DNA monomer, 12 kDa DNA
DNA dC->dU-editing enzyme APOBEC-3G monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
118 |
nm3 |
|
|