|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNU4_fit1_model1.png)
|
| Sample: |
Di[3-deoxy-D-manno-octulosonyl]-lipid A (ammonium salt), unidentified lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNV4_fit1_model1.png)
|
| Sample: |
Di[3-deoxy-D-manno-octulosonyl]-lipid A (ammonium salt) plus WLBU2, unidentified lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNW4_fit1_model1.png)
|
| Sample: |
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(10-rac-glycerol) sodium salt, 10,30-bis-[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (7:2:1), lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2021 Jun 23
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNX4_fit1_model1.png)
|
| Sample: |
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(10-rac-glycerol) sodium salt, 10,30-bis-[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol sodium salt, WLBU2, synthetic construct lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNY4_fit1_model1.png)
|
| Sample: |
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(10-rac-glycerol), 1,2-dioleoyl-3-trimethylammonium-propane, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 10,30-bis-[1,2-dioleoyl-sn-glycero-3-phos, lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNZ4_fit1_model1.png)
|
| Sample: |
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(10-rac-glycerol), 1,2-dioleoyl-3-trimethylammonium-propane, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 10,30-bis-[1,2-dioleoyl-sn-glycero-3-phos, synthetic construct lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDN25_fit1_model1.png)
|
| Sample: |
Lipopolysaccharide, Pseudomonas aeruginosa lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Jun 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDN35_fit1_model1.png)
|
| Sample: |
Lipopolysaccharide plus WLBU2 dimer, Pseudomonas aeruginosa lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Jun 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
|
| Sample: |
Histone deacetylase 6 monomer, 131 kDa Homo sapiens protein
|
| Buffer: |
30 mM HEPES, 140 mM NaCl, 10 mM KCl, 0.25 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 10
|
In-solution structure and oligomerization of human histone deacetylase 6 - an integrative approach.
FEBS J (2022)
Shukla S, Komarek J, Novakova Z, Nedvedova J, Ustinova K, Vankova P, Kadek A, Uetrecht C, Mertens H, Barinka C
|
| RgGuinier |
7.0 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
316 |
nm3 |
|
|
|
|
|
|
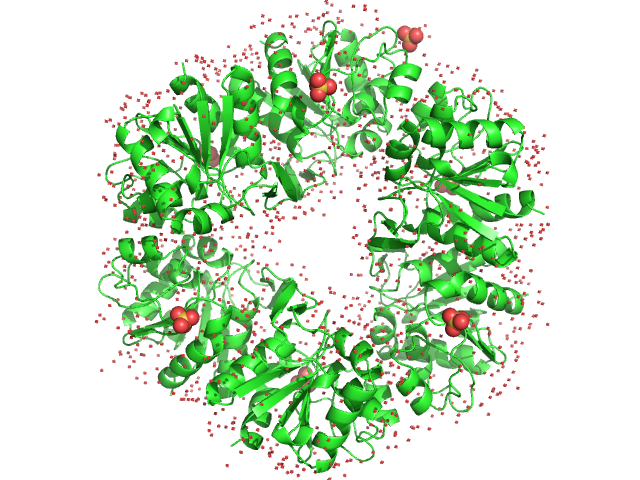
|
| Sample: |
Deglycase PH1704 hexamer, 112 kDa Pyrococcus horikoshii (strain … protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Jun 7
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
140 |
nm3 |
|
|