|
|
|
|
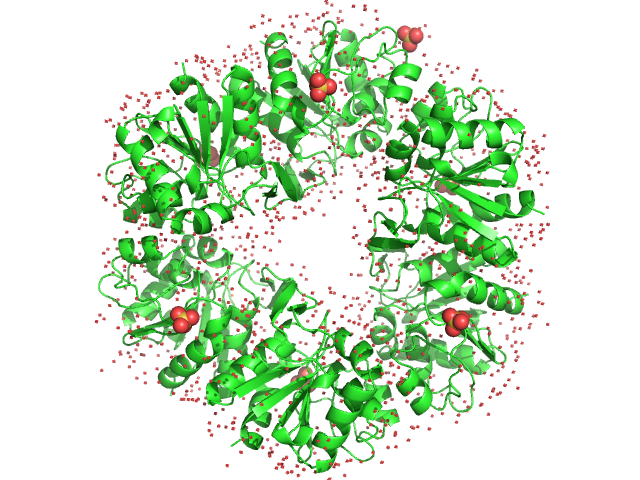
|
| Sample: |
Deglycase PH1704 hexamer, 112 kDa Pyrococcus horikoshii (strain … protein
|
| Buffer: |
20 mM Tris pH 7.5 and 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
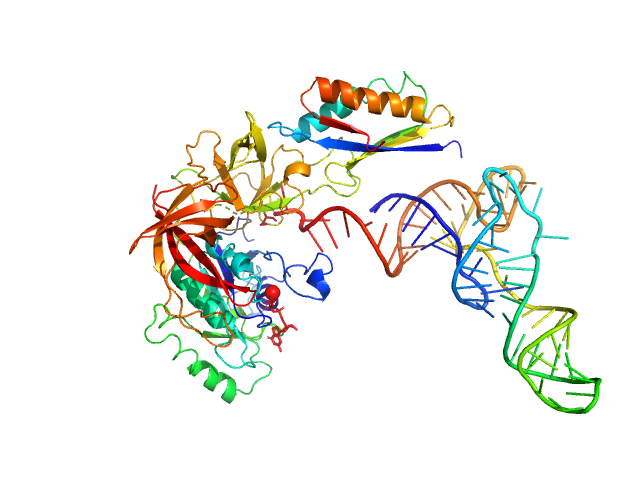
|
| Sample: |
Translation initiation factor 2 subunit gamma monomer, 46 kDa Saccharolobus solfataricus (strain … protein
Translation initiation factor 2 subunit alpha monomer, 10 kDa Saccharolobus solfataricus (strain … protein
Transfer RNA monomer, 23 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS- NaOH pH 6.7, 200 mM NaCl, 5 mM MgCl 2, 1 mM GDPNP, pH: 6.7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
C-phycocyanin alpha subunit trimer, 53 kDa Arthrospira platensis protein
C-phycocyanin beta subunit trimer, 54 kDa Arthrospira platensis protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, pH: 7.6 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Apr 14
|
Anti-Stokes fluorescence excitation reveals conformational mobility of the C-phycocyanin chromophores
Structural Dynamics 9(5):054701 (2022)
Tsoraev G, Protasova E, Klimanova E, Ryzhykau Y, Kuklin A, Semenov Y, Ge B, Li W, Qin S, Friedrich T, Sluchanko N, Maksimov E
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
C-phycocyanin alpha subunit monomer, 18 kDa Arthrospira platensis protein
C-phycocyanin beta subunit monomer, 18 kDa Arthrospira platensis protein
|
| Buffer: |
8 M urea, 30 mM βME, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Apr 14
|
Anti-Stokes fluorescence excitation reveals conformational mobility of the C-phycocyanin chromophores
Structural Dynamics 9(5):054701 (2022)
Tsoraev G, Protasova E, Klimanova E, Ryzhykau Y, Kuklin A, Semenov Y, Ge B, Li W, Qin S, Friedrich T, Sluchanko N, Maksimov E
|
| RgGuinier |
6.6 |
nm |
| Dmax |
23.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Toll-like receptor dimer, 175 kDa Aedes aegypti protein
|
| Buffer: |
50 mM NaCl, 50 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Aug 2
|
Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun 13(1):5110 (2022)
Saucereau Y, Wilson TH, Tang MCK, Moncrieffe MC, Hardwick SW, Chirgadze DY, Soares SG, Marcaida MJ, Gay NJ, Gangloff M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Toll-like receptor dimer, 175 kDa Aedes aegypti protein
AAEL013433-PA (Cystine-knot_cytokine) dimer, 26 kDa Aedes aegypti protein
|
| Buffer: |
50 mM NaCl, 50 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Aug 2
|
Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun 13(1):5110 (2022)
Saucereau Y, Wilson TH, Tang MCK, Moncrieffe MC, Hardwick SW, Chirgadze DY, Soares SG, Marcaida MJ, Gay NJ, Gangloff M
|
| RgGuinier |
5.5 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
567 |
nm3 |
|
|
|
|
|
|
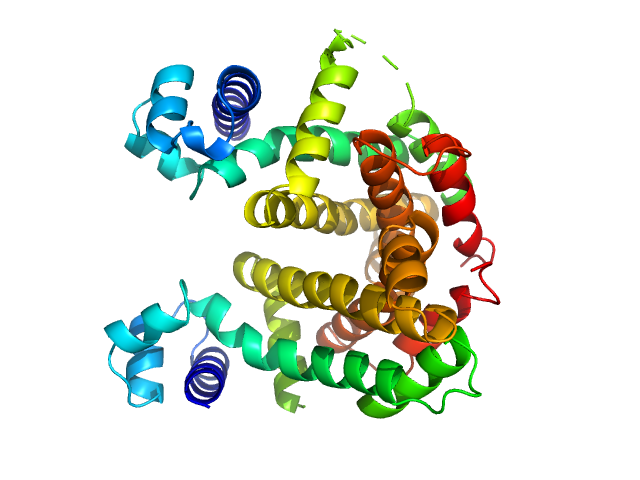
|
| Sample: |
Putative tetR-family transcriptional regulator dimer, 47 kDa Streptomyces coelicolor (strain … protein
|
| Buffer: |
20 mM Tris/HCl, 400 mM NaCl, 50 mM imidazole, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 May 2
|
Crystal Structures of Free and Ligand-Bound Forms of the TetR/AcrR-Like Regulator SCO3201 from Streptomyces coelicolor Suggest a Novel Allosteric Mechanism.
FEBS J (2022)
Werten S, Waack P, Palm GJ, Virolle MJ, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
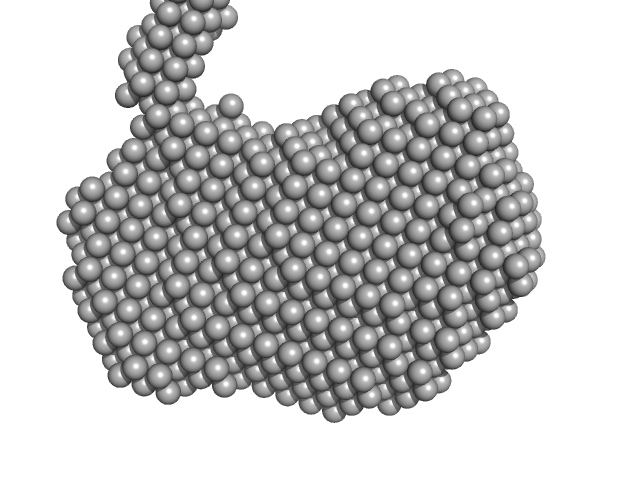
|
| Sample: |
LipAMS8 monomer, 50 kDa Pseudomonas sp. AMS8 protein
|
| Buffer: |
50 mM Tris HCl, 5 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 Mar 6
|
Structural interpretations of a flexible cold-active AMS8 lipase by combining small-angle X-ray scattering and molecular dynamics simulation (SAXS-MD).
Int J Biol Macromol (2022)
Yaacob N, Kamonsutthipaijit N, Soontaranon S, Leow TC, Rahman RNZRA, Ali MSM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Homo sapiens RNA component of 7SK nuclear ribonucleoprotein (RN7SK), small nuclear RNA monomer, 18 kDa Homo sapiens RNA
Protein Tat monomer, 2 kDa Human immunodeficiency virus … protein
|
| Buffer: |
10 mM phosphate, 70 mM NaCl, 0.1 mM EDTA, pH: 5.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 27
|
A structure-based mechanism for displacement of the HEXIM adapter from 7SK small nuclear RNA.
Commun Biol 5(1):819 (2022)
Pham VV, Gao M, Meagher JL, Smith JL, D'Souza VM
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Homo sapiens RNA component of 7SK nuclear ribonucleoprotein (RN7SK), small nuclear RNA monomer, 18 kDa Homo sapiens RNA
Protein HEXIM1 monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 70 mM NaCl, 0.1 mM EDTA, pH: 5.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 27
|
A structure-based mechanism for displacement of the HEXIM adapter from 7SK small nuclear RNA.
Commun Biol 5(1):819 (2022)
Pham VV, Gao M, Meagher JL, Smith JL, D'Souza VM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
26 |
nm3 |
|
|