|
|
|
|
|
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Estrogen receptor (mutant S118A) monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Estrogen receptor (mutant S118D) monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 0.05 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jul 17
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
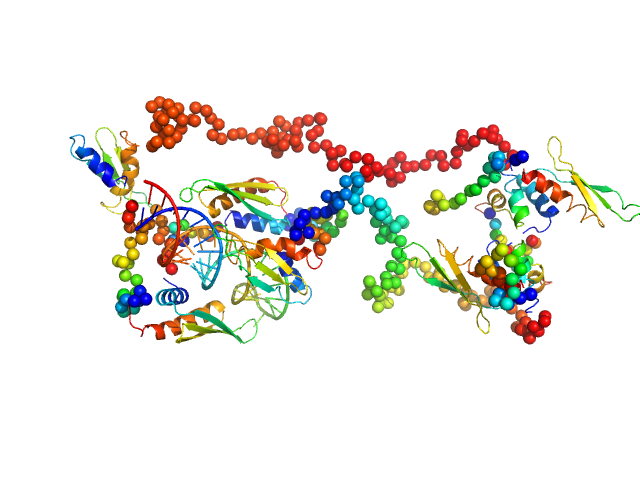
| Sample: |
Fc fragment of IgG binding protein dimer, 552 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 10 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 20
|
The structure of FCGBP is formed as a disulfide-mediated homodimer between its C-terminal domains.
FEBS J (2025)
Ehrencrona E, Gallego P, Trillo-Muyo S, Garcia-Bonete MJ, Recktenwald CV, Hansson GC, Johansson MEV
|
| RgGuinier |
10.0 |
nm |
| Dmax |
42.0 |
nm |
| VolumePorod |
1462 |
nm3 |
|
|
|
|
|
|
|
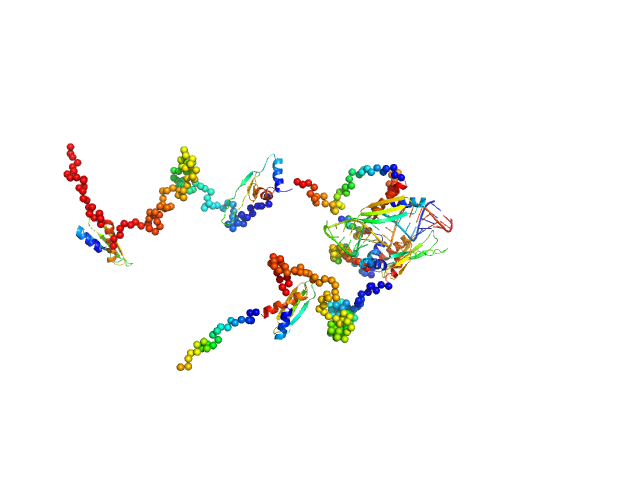
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 (Δ1-177) dimer, 89 kDa Homo sapiens protein
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2020 May 4
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
Double-stranded RNA-binding protein Staufen homolog 1 with truncated RNA-binding domain 2 and truncated Staufen-swapping (ΔSSM) dimer, 81 kDa Homo sapiens protein
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2024 Jan 12
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
5.5 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
|
|
|
|
|
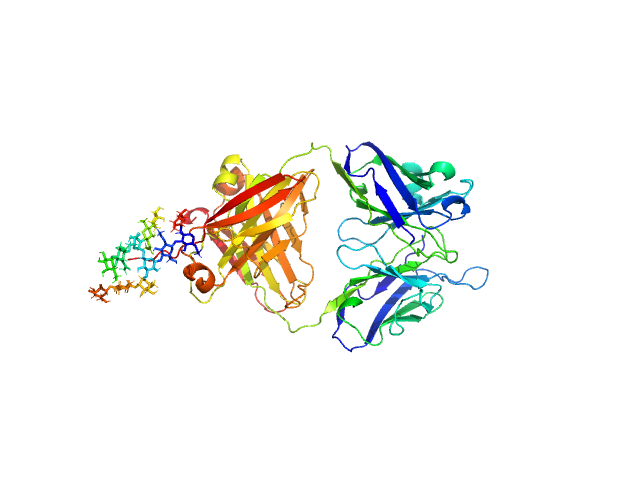
| Sample: |
IgM Mannitou Fab Heavy Chain monomer, 26 kDa Mus musculus protein
IgM Mannitou Fab Light Chain monomer, 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Jun 5
|
Small-angle X-ray scattering of engineered antigen-binding fragments: the case of glycosylated Fab from the Mannitou IgM antibody.
Acta Crystallogr F Struct Biol Commun (2025)
Semwal S, Karamolegkou M, Flament S, Raouraoua N, Verstraete K, Thureau A, Wien F, Bray F, Savvides SN, Bouckaert J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
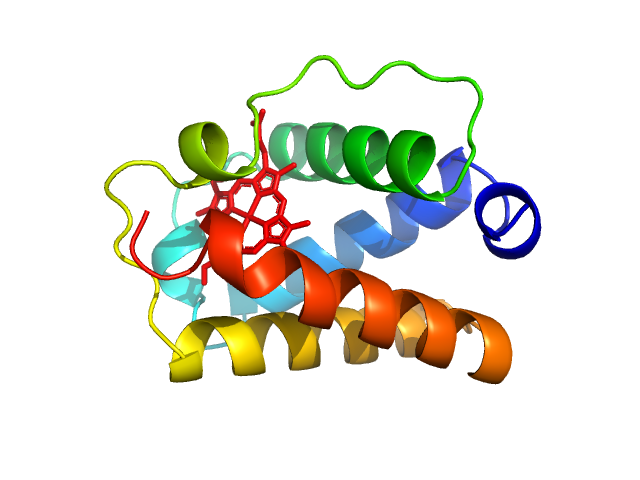
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 May 1
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|