|
|
|
|
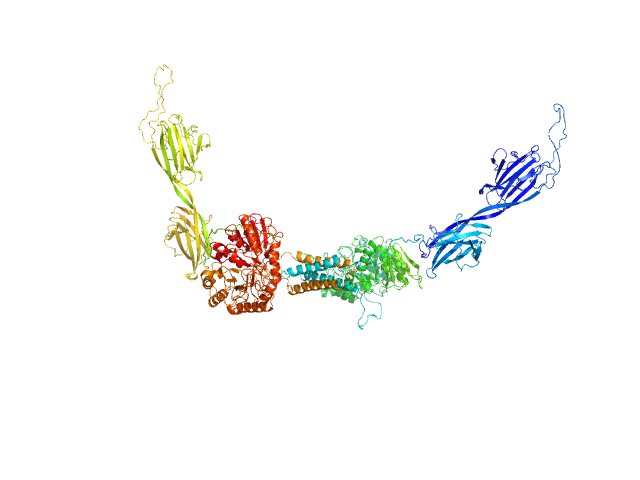
|
| Sample: |
Alpha-amylase 3, chloroplastic dimer, 187 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
444 |
nm3 |
|
|
|
|
|
|
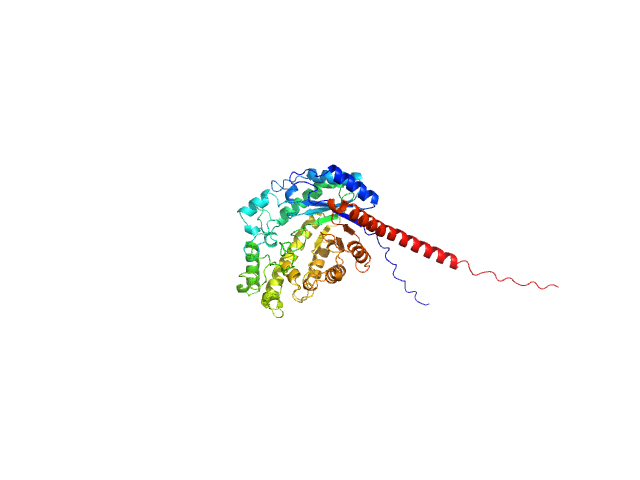
|
| Sample: |
Inactive beta-amylase 9 monomer, 50 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
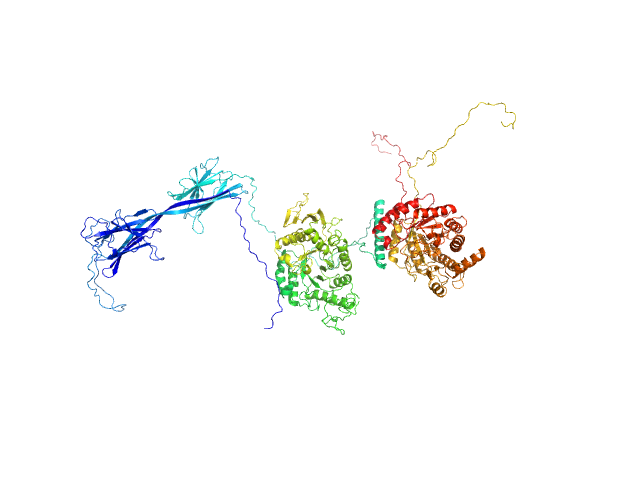
|
| Sample: |
Inactive beta-amylase 9 monomer, 50 kDa Arabidopsis thaliana protein
Alpha-amylase 3, chloroplastic monomer, 94 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
5.0 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|
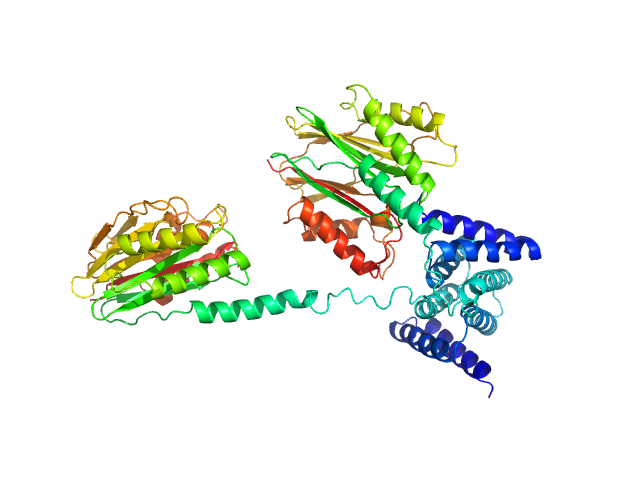
|
| Sample: |
Phosphoserine phosphatase RsbU dimer, 77 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 24
|
A General Mechanism for Initiating the General Stress Response in Bacteria.
bioRxiv (2025)
Baral R, Ho K, Kumar RP, Hopkins JB, Watkins MB, LaRussa S, Caban-Penix S, Calderone LA, Bradshaw N
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/threonine-protein kinase RsbT dimer, 29 kDa Bacillus subtilis (strain … protein
Phosphoserine phosphatase RsbU (Q94L) dimer, 77 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 24
|
A General Mechanism for Initiating the General Stress Response in Bacteria.
bioRxiv (2025)
Baral R, Ho K, Kumar RP, Hopkins JB, Watkins MB, LaRussa S, Caban-Penix S, Calderone LA, Bradshaw N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
119 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 dimer, 12 kDa Homo sapiens protein
Apoptosis-inducing factor 1, mitochondrial dimer, 113 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 28
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
289 |
nm3 |
|
|