|
|
|
|
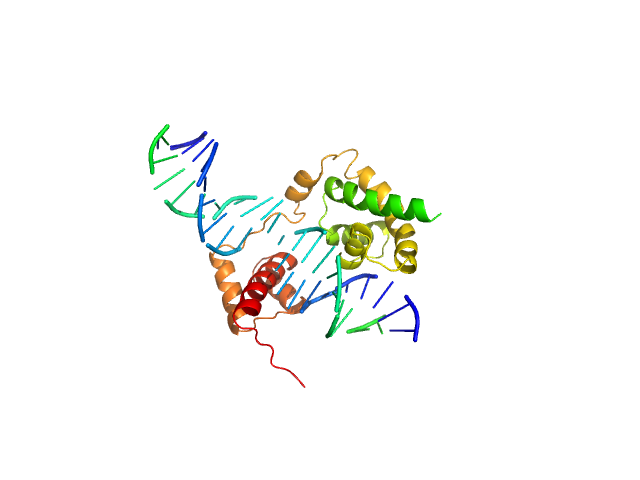
|
| Sample: |
POU domain, class 3, transcription factor 2 monomer, 19 kDa Homo sapiens protein
Rat CRH DNA monomer, 15 kDa Rattus norvegicus DNA
|
| Buffer: |
50 mM Tris, 0.4 M NaCl, 2% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2003 Oct 9
|
Fine-tuning of intrinsic N-Oct-3 POU domain allostery by regulatory DNA targets
Nucleic Acids Research 35(13):4420-4432 (2007)
Alazard R, Mourey L, Ebel C, Konarev P, Petoukhov M, Svergun D, Erard M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
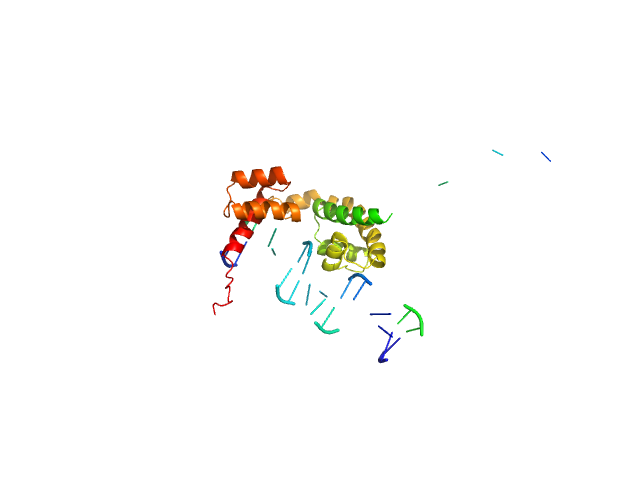
|
| Sample: |
POU domain, class 3, transcription factor 2 monomer, 19 kDa Homo sapiens protein
Human DR-alpha DNA monomer, 15 kDa Homo sapiens DNA
|
| Buffer: |
50 mM Tris, 0.4 M NaCl, 2% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 May 26
|
Fine-tuning of intrinsic N-Oct-3 POU domain allostery by regulatory DNA targets
Nucleic Acids Research 35(13):4420-4432 (2007)
Alazard R, Mourey L, Ebel C, Konarev P, Petoukhov M, Svergun D, Erard M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Filamin C 23-24 dimer, 40 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jun 17
|
Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23-24 dimer.
J Mol Biol 368(4):1011-23 (2007)
Sjekloća L, Pudas R, Sjöblom B, Konarev P, Carugo O, Rybin V, Kiema TR, Svergun D, Ylänne J, Djinović Carugo K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
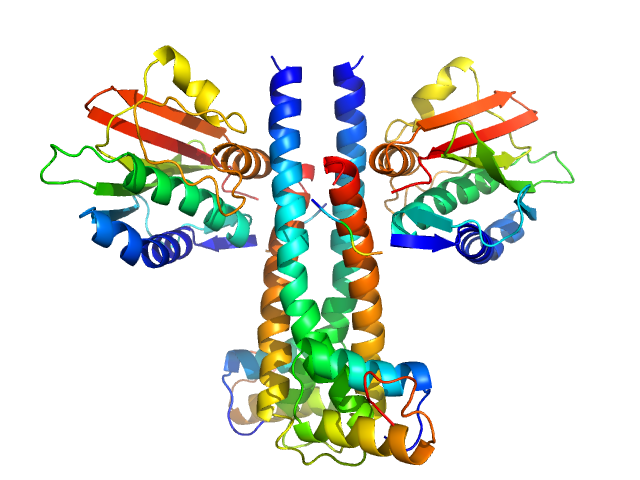
|
| Sample: |
Sporulation kinase A dimer, 51 kDa Bacillus subtilis protein
Sporulation inhibitor sda dimer, 11 kDa Bacillus subtilis protein
|
| Buffer: |
50mM Tris, 200mM NaCl, 150mM Imidazole, pH: 8.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Nov 16
|
The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition.
J Mol Biol 368(2):407-20 (2007)
Whitten AE, Jacques DA, Hammouda B, Hanley T, King GF, Guss JM, Trewhella J, Langley DB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
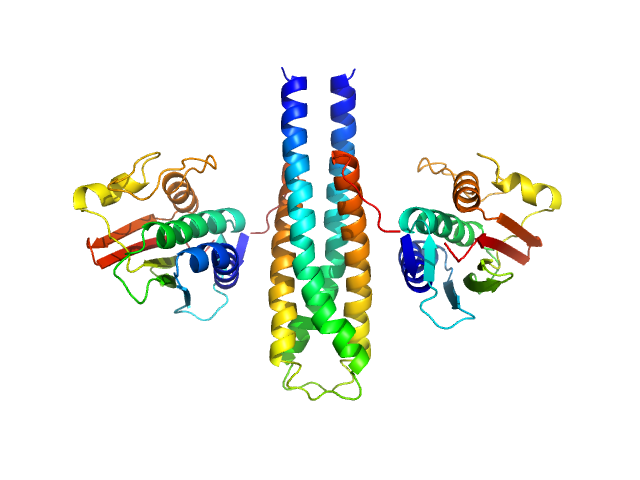
|
| Sample: |
Sporulation kinase A dimer, 51 kDa Bacillus subtilis protein
|
| Buffer: |
50mM Tris, 200mM NaCl, 150mM Imidazole, pH: 8.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Mar 18
|
The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition.
J Mol Biol 368(2):407-20 (2007)
Whitten AE, Jacques DA, Hammouda B, Hanley T, King GF, Guss JM, Trewhella J, Langley DB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
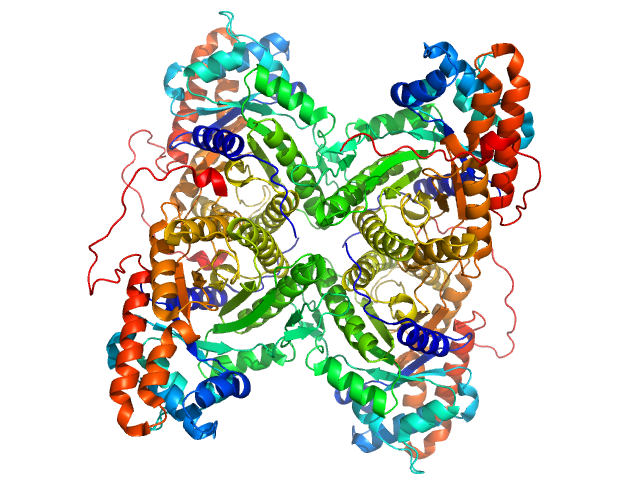
|
| Sample: |
Fructose-bisphosphate aldolase A tetramer, 157 kDa Oryctolagus cuniculus protein
|
| Buffer: |
100 mM Tris/HCl 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 19
|
Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering
Journal of Applied Crystallography 40(s1):s245-s249 (2007)
Mylonas E, Svergun D
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.5 |
nm |
|
|
|
|
|
|
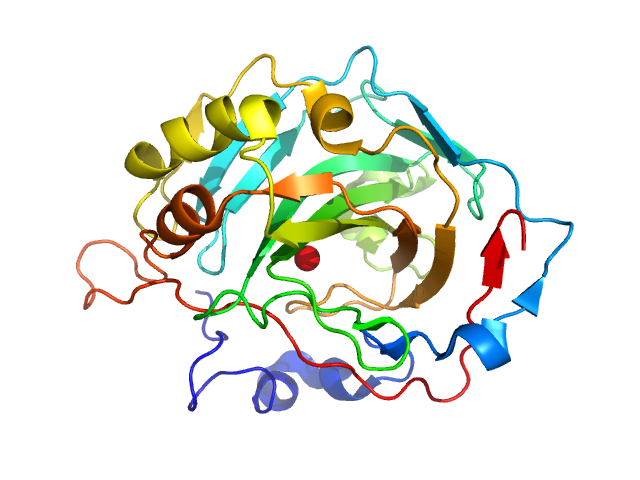
|
| Sample: |
Carbonic anhydrase 2 monomer, 29 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris/HCl 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 19
|
Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering
Journal of Applied Crystallography 40(s1):s245-s249 (2007)
Mylonas E, Svergun D
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Thyroglobulin dimer, 606 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris/HCl 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 19
|
Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering
Journal of Applied Crystallography 40(s1):s245-s249 (2007)
Mylonas E, Svergun D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
24.0 |
nm |
|
|
|
|
|
|
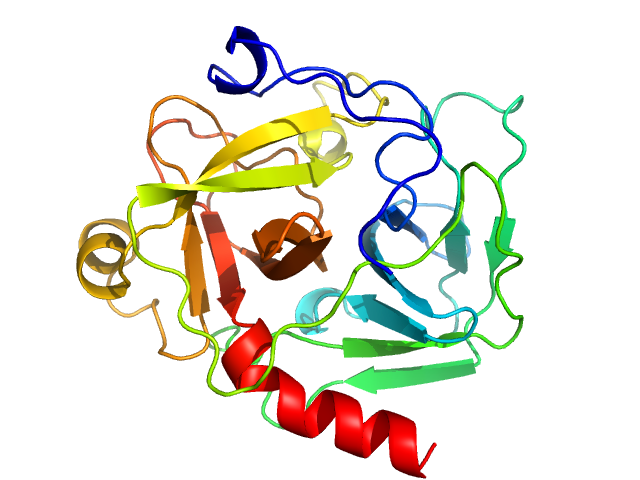
|
| Sample: |
Chymotrypsinogen A monomer, 26 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris/HCl 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 19
|
Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering
Journal of Applied Crystallography 40(s1):s245-s249 (2007)
Mylonas E, Svergun D
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Major prion protein 24-mer, 664 kDa Homo sapiens protein
|
| Buffer: |
5 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jun 2
|
Structural characterization of beta-sheeted oligomers formed on the pathway of oxidative prion protein aggregation in vitro.
J Struct Biol 157(2):308-20 (2007)
Redecke L, von Bergen M, Clos J, Konarev PV, Svergun DI, Fittschen UE, Broekaert JA, Bruns O, Georgieva D, Mandelkow E, Genov N, Betzel C
|
| RgGuinier |
9.8 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
3320 |
nm3 |
|
|