|
|
|
|
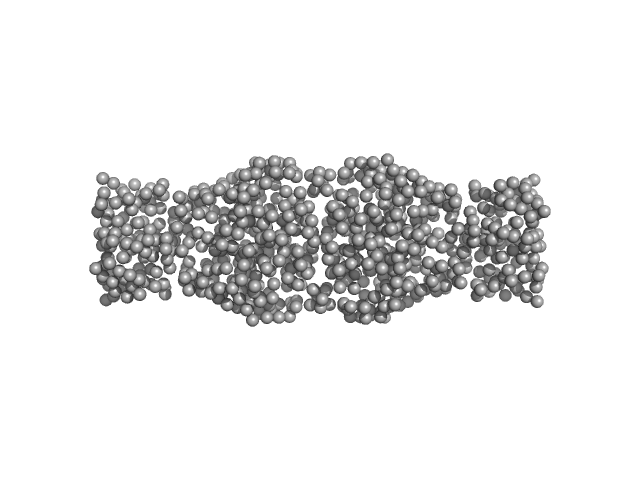
|
| Sample: |
Human hemoglobin conjugated with six-seven copies of 5-kDa PEG dimer, 62 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Human hemoglobin conjugated with two copies of 5-kDa PEG dimer, 62 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
|
|
|
|
|
|

|
| Sample: |
Human hemoglobin conjugated with two copies of 5-kDa PEG dimer, 62 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.0 |
nm |
|
|
|
|
|
|
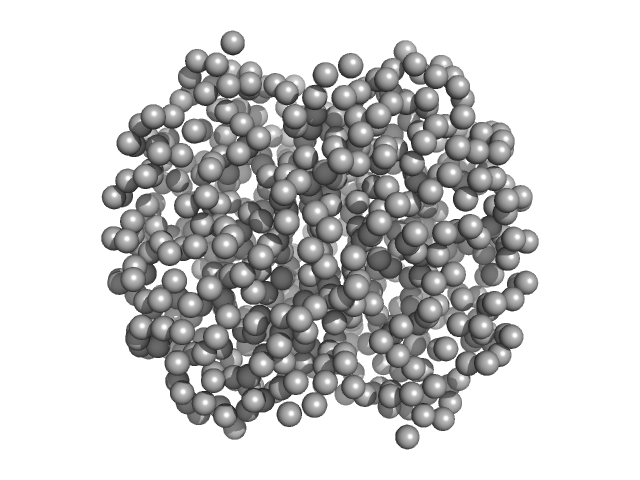
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
13.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
|
|
|
|
|
|
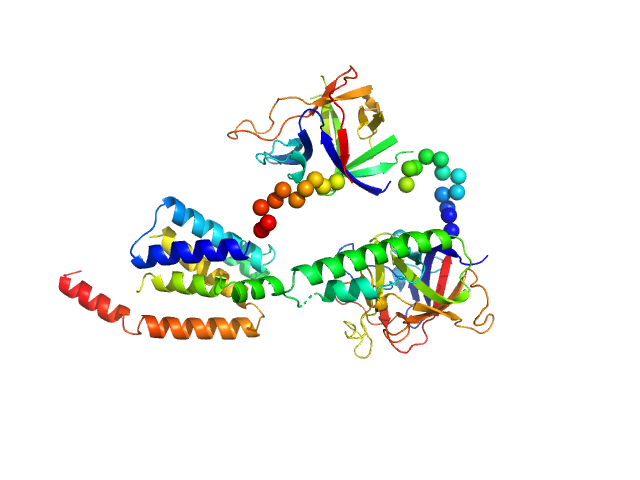
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 5 mM EGTA, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
|
|
|
|
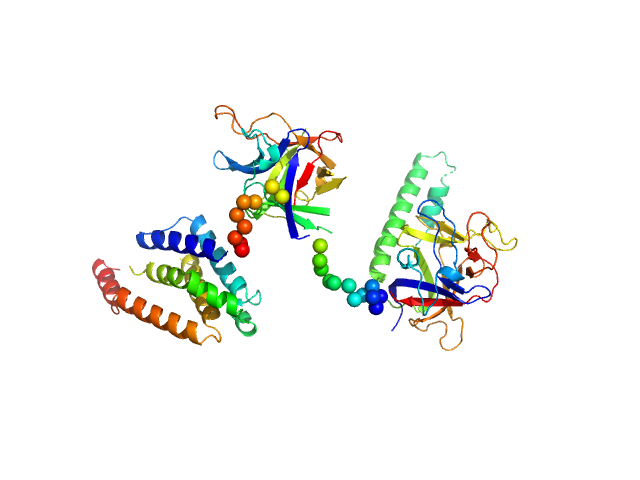
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
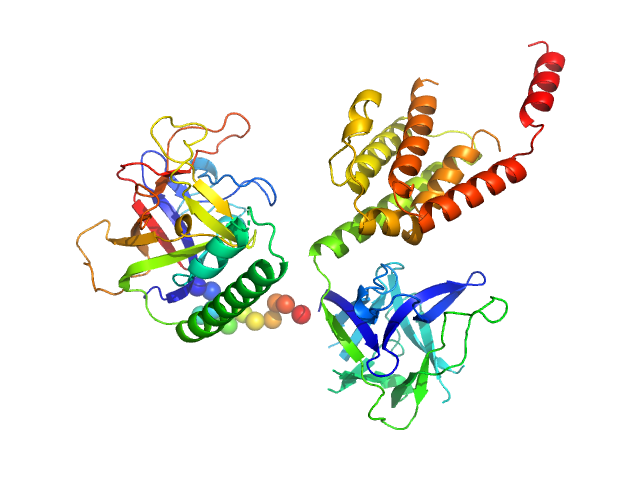
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 5 mM EGTA, 0.25 mM IP3, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
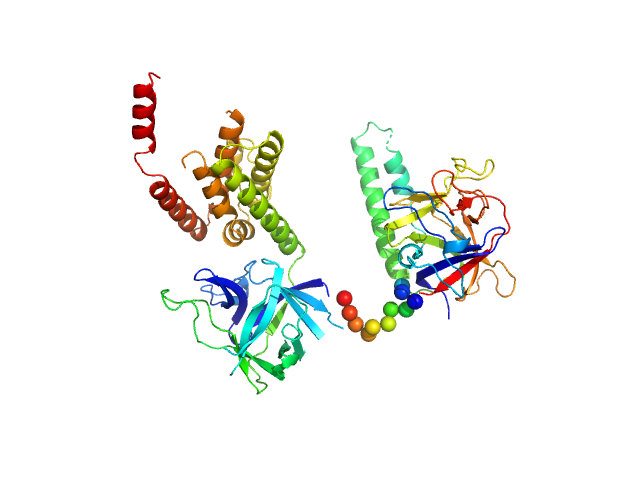
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 10 mM CaCl2, 0.25 mM IP3, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
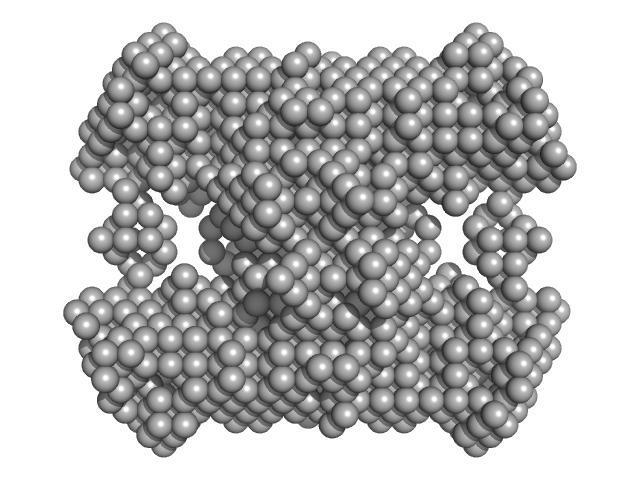
| Sample: |
N-acetylglucosamine kinase 1 tetramer, 219 kDa Candida albicans (strain … protein
|
| Buffer: |
0.2 M magnesium acetate, 0.1 M sodium cacodylate, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Apr 28
|
The crystal and solution studies of glucosamine-6-phosphate synthase from Candida albicans.
J Mol Biol 372(3):672-88 (2007)
Raczynska J, Olchowy J, Konariev PV, Svergun DI, Milewski S, Rypniewski W
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
421 |
nm3 |
|
|