|
|
|
|
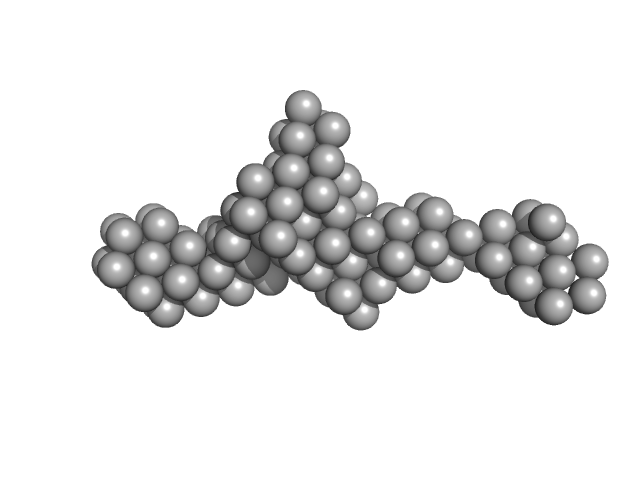
|
| Sample: |
Z-DNA-binding protein 1 dimer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
ZBP1 condensate formation synergizes Z-NAs recognition and signal transduction.
Cell Death Dis 15(7):487 (2024)
Xie F, Wu D, Huang J, Liu X, Shen Y, Huang J, Su Z, Li J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
5_SL1 monomer, 9 kDa Severe acute respiratory … RNA
|
| Buffer: |
50 mM BisTris, 25 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 1
|
The 5'-terminal stem-loop RNA element of SARS-CoV-2 features highly dynamic structural elements that are sensitive to differences in cellular pH.
Nucleic Acids Res (2024)
Toews S, Wacker A, Faison EM, Duchardt-Ferner E, Richter C, Mathieu D, Bottaro S, Zhang Q, Schwalbe H
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
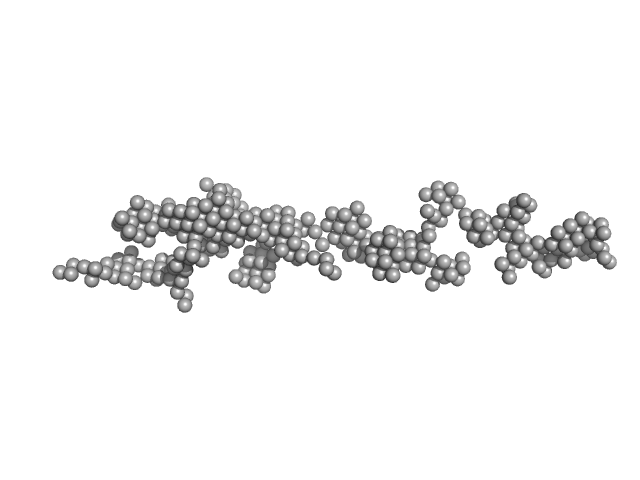
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 11
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
5.8 |
nm |
| Dmax |
42.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 13
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
6.9 |
nm |
| Dmax |
60.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 31
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
2.5 |
nm |
| Dmax |
6.5 |
nm |
|
|
|
|
|
|
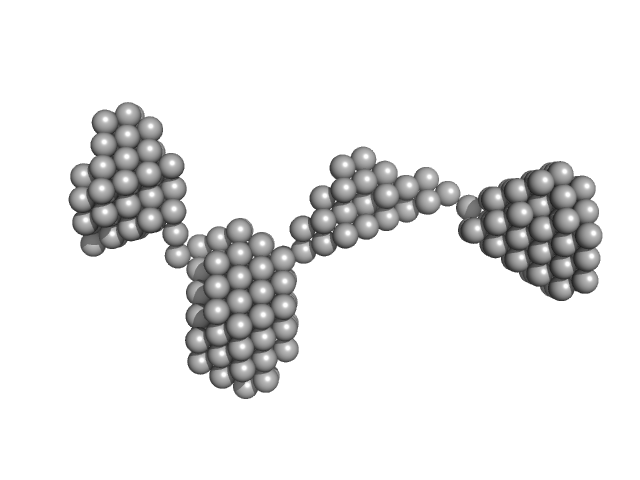
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 1
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.7 |
nm |
| Dmax |
9.1 |
nm |
|
|
|
|
|
|
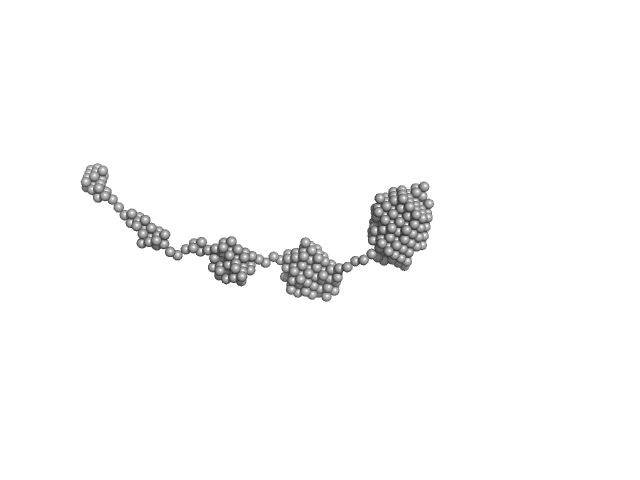
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 1
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 2
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Endonuclease 8 2 dimer, 108 kDa Mycobacterium tuberculosis (strain … protein
Single-stranded DNA-binding protein octamer, 139 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Jul 10
|
Identification and characterization of MtbNei2-SSB forms novel pre-replicative BER complex in Mycobacterium tuberculosis.
Jyoti Vishwakarma
|
| RgGuinier |
5.5 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
734 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Feb 15
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
253 |
nm3 |
|
|