|
|
|
|
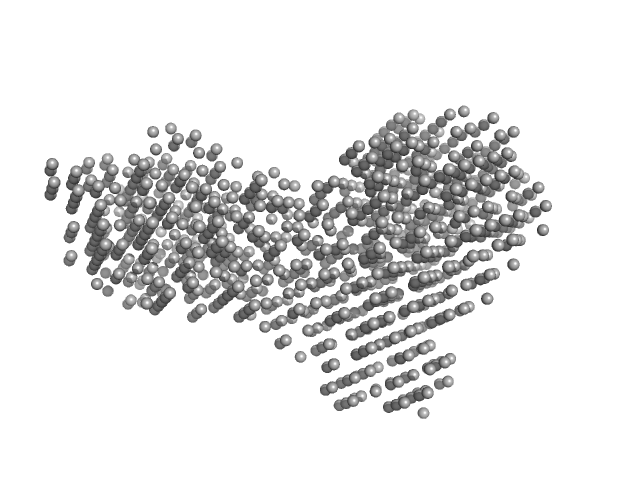
|
| Sample: |
Beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jun 4
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
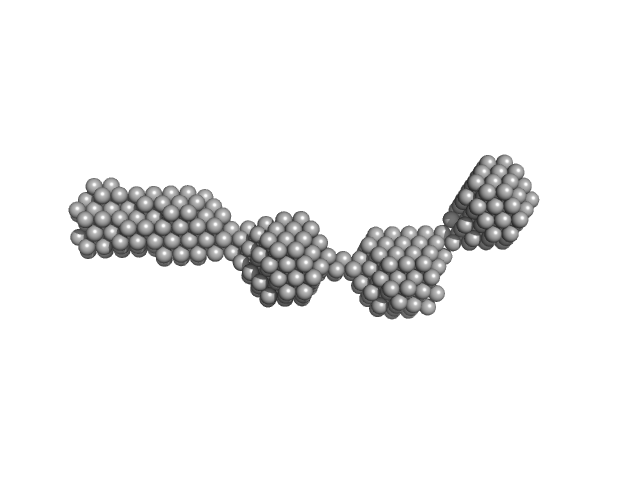
|
| Sample: |
Nuclear fusion protein BIK1 dimer, 106 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 500 mM NaCl, 2% glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jul 13
|
Phase separation of a microtubule plus-end tracking protein into a fluid fractal network
(2024)
Czub M, Uliana F, Grubić T, Padeste C, Rosowski K, Dufresne E, Menzel A, Vakonakis I, Gasser U, Steinmetz M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
467 |
nm3 |
|
|
|
|
|
|
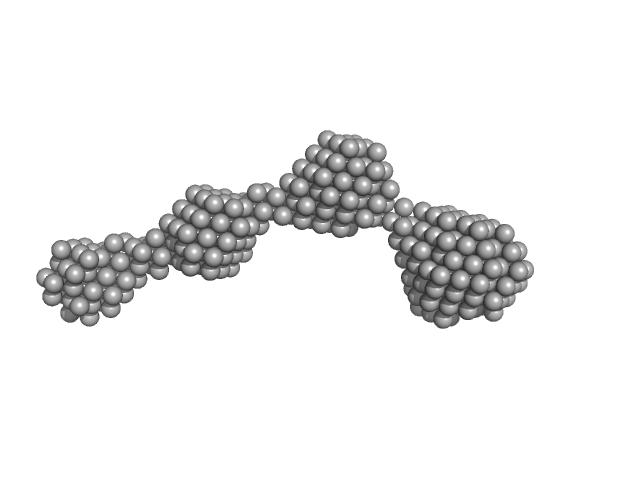
|
| Sample: |
Nuclear fusion protein BIK1 dimer, 55 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 500 mM NaCl, 2% glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jul 13
|
Phase separation of a microtubule plus-end tracking protein into a fluid fractal network
(2024)
Czub M, Uliana F, Grubić T, Padeste C, Rosowski K, Dufresne E, Menzel A, Vakonakis I, Gasser U, Steinmetz M
|
| RgGuinier |
8.0 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
282 |
nm3 |
|
|
|
|
|
|
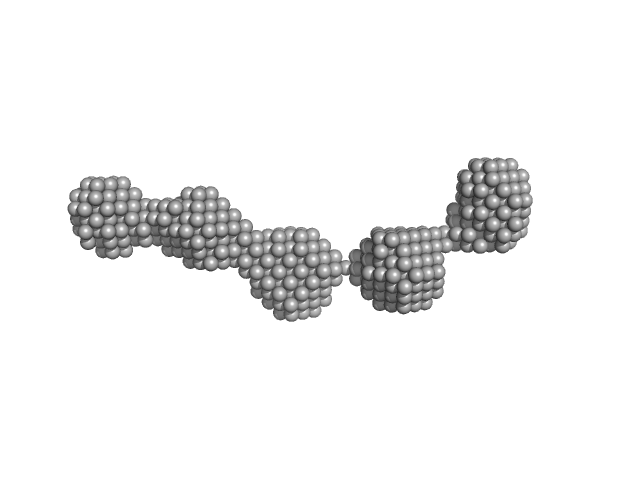
|
| Sample: |
Nuclear fusion protein BIK1 dimer, 105 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 500 mM NaCl, 2% glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jul 13
|
Phase separation of a microtubule plus-end tracking protein into a fluid fractal network
(2024)
Czub M, Uliana F, Grubić T, Padeste C, Rosowski K, Dufresne E, Menzel A, Vakonakis I, Gasser U, Steinmetz M
|
| RgGuinier |
9.4 |
nm |
| Dmax |
29.4 |
nm |
| VolumePorod |
632 |
nm3 |
|
|
|
|
|
|
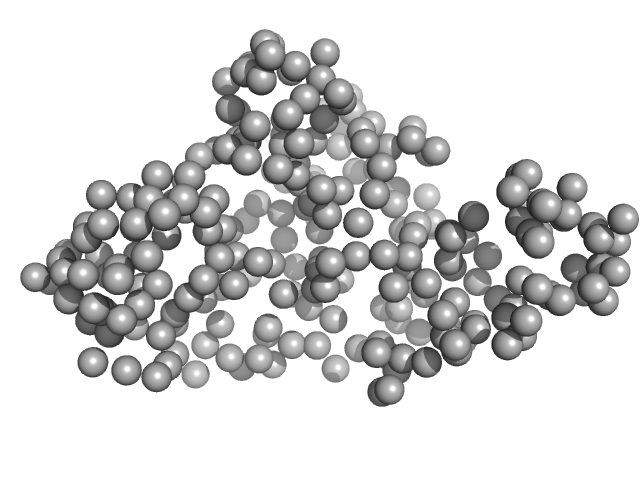
|
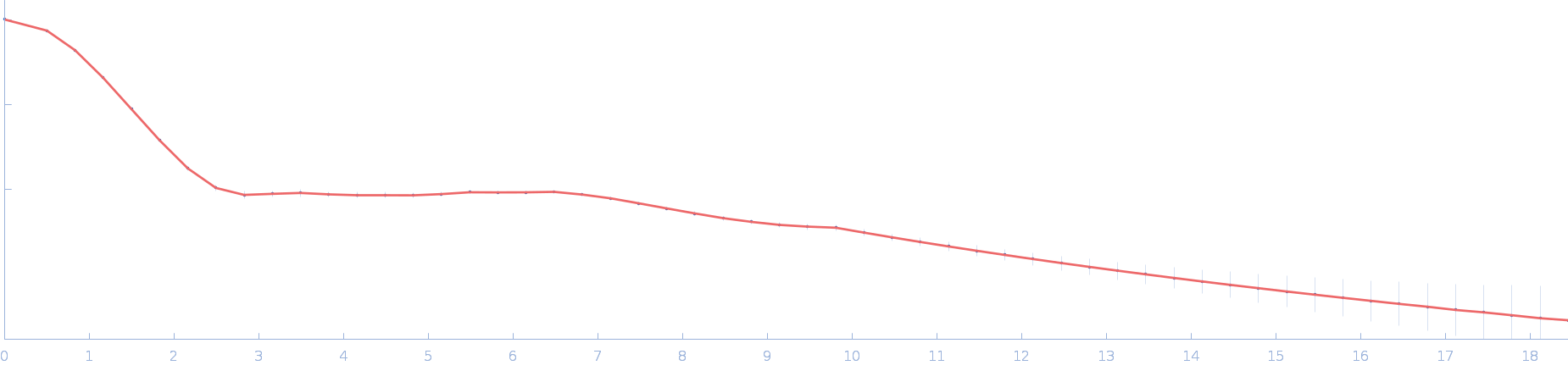
| Sample: |
RTX toxin transporter monomer, 28 kDa Kingella kingae ATCC … protein
|
| Buffer: |
100 mM HEPES, 10 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
Type 1 secretion necessitates a tight interplay between all domains of the ABC transporter
Scientific Reports 14(1) (2024)
Anlauf M, Bilsing F, Reiners J, Spitz O, Hachani E, Smits S, Schmitt L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
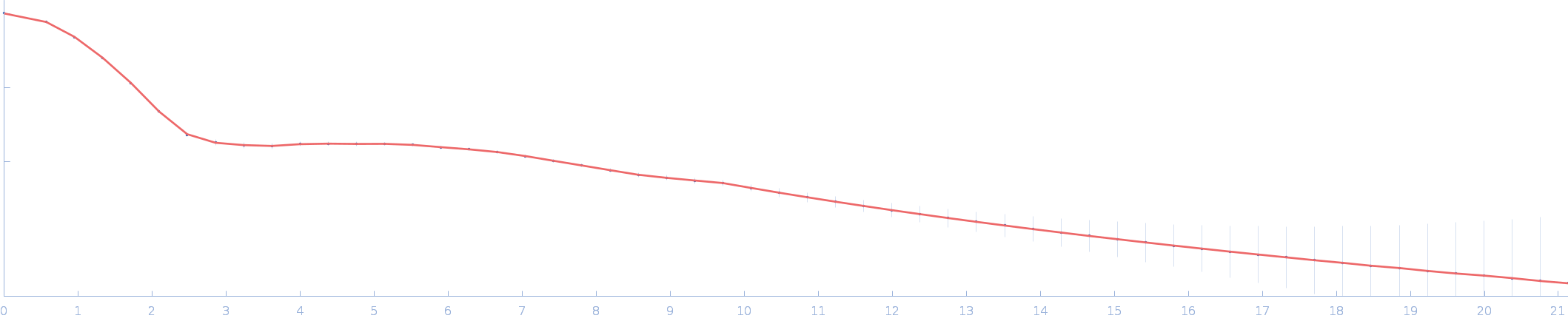
| Sample: |
Ssr1698 protein (H79A:R90A) dimer, 22 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Mar 30
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
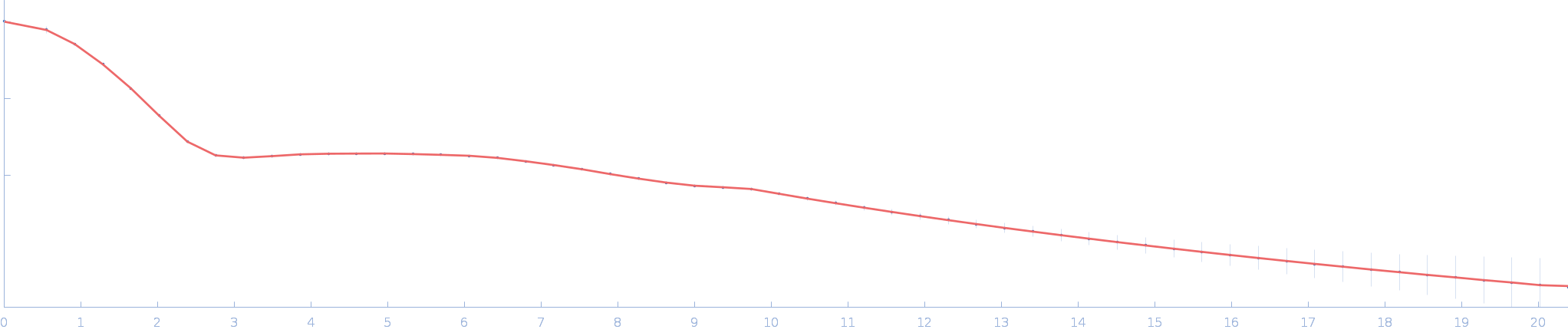
| Sample: |
Ssr1698 protein (H79A:R90A) monomer, 11 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Mar 30
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ssr1698 protein (H21A) monomer, 11 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Jun 10
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
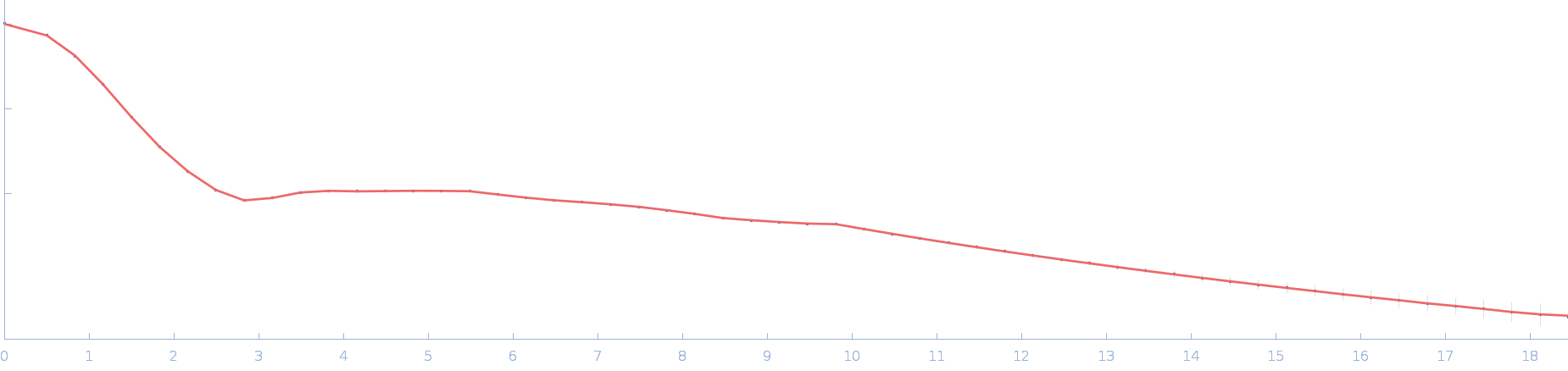
| Sample: |
Ssr1698 protein dimer, 23 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Sep 27
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
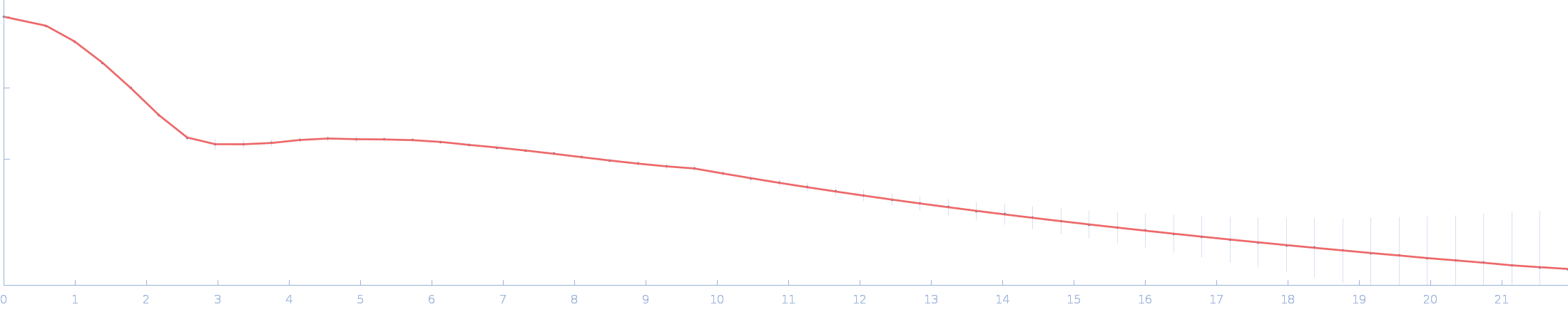
| Sample: |
Ssr1698 protein monomer, 11 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2021 Dec 12
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.3 |
nm |
| VolumePorod |
12 |
nm3 |
|
|