|
|
|
|
|
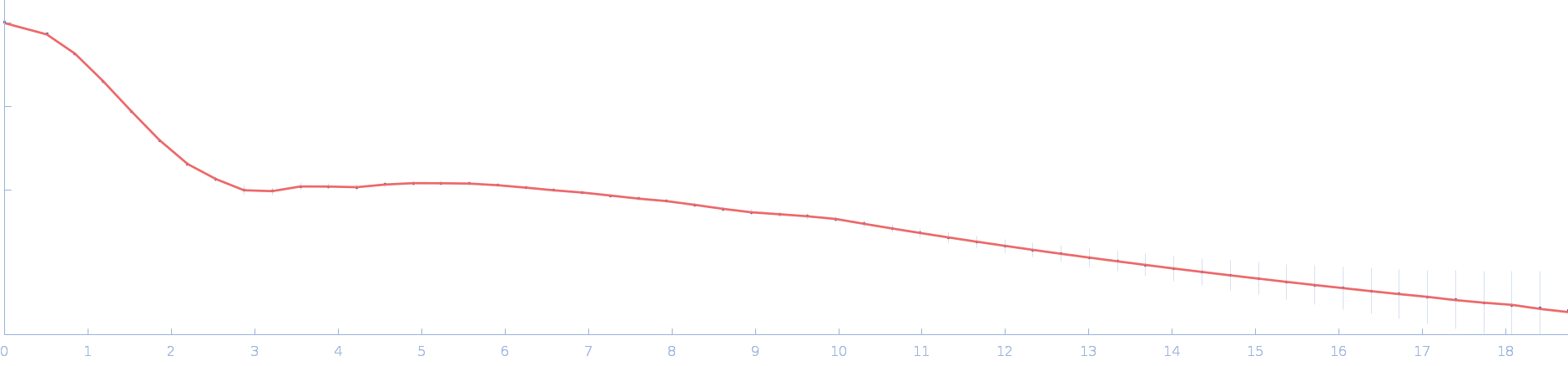
| Sample: |
Ssr1698 protein dimer, 23 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Jun 13
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
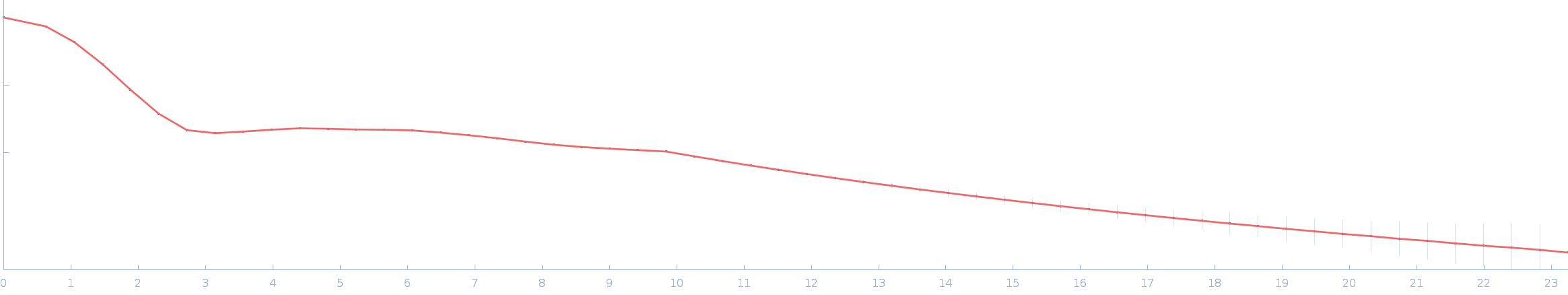
| Sample: |
Ssr1698 protein monomer, 11 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Sep 27
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
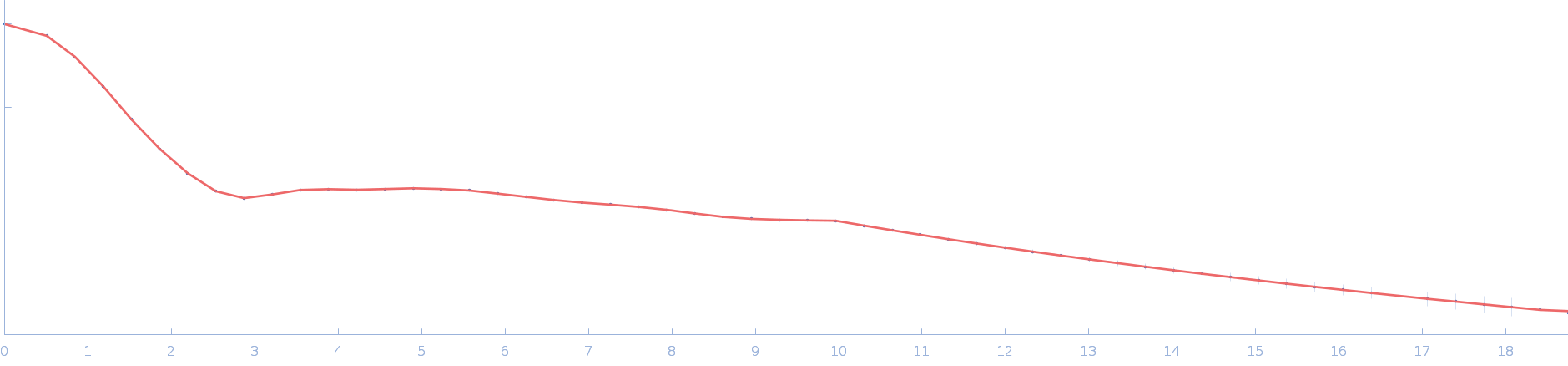
| Sample: |
Ssr1698 protein dimer, 23 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Sep 27
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
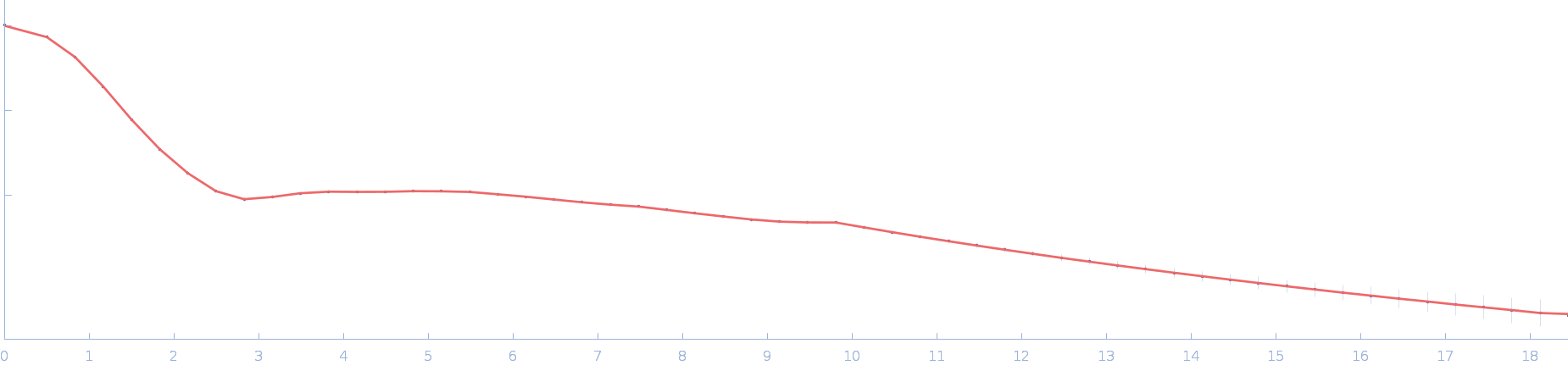
| Sample: |
Ssr1698 protein dimer, 23 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Jun 13
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
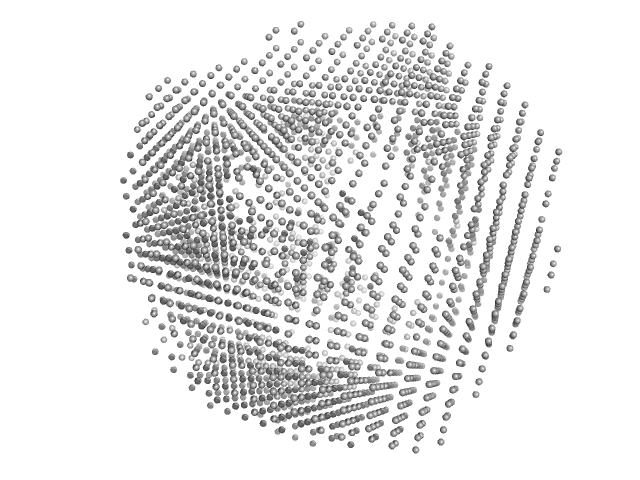
|
| Sample: |
N-terminal split fragment of a single subunit of Truncated Icosahedral Protein composed of 60-mer fusion proteins, 360 kDa synthetic construct protein
C-terminal split fragment of a single subunit of Truncated Isosahedral Protein composed of 60-mer fusion proteins, 715 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 5%(v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2023 Nov 27
|
Fusion then fission: splitting and reassembly of an artificial fusion-protein nanocage.
Chem Commun (Camb) (2024)
Ohara N, Kawakami N, Arai R, Adachi N, Ikeda A, Senda T, Miyamoto K
|
| RgGuinier |
9.2 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
3480 |
nm3 |
|
|
|
|
|
|
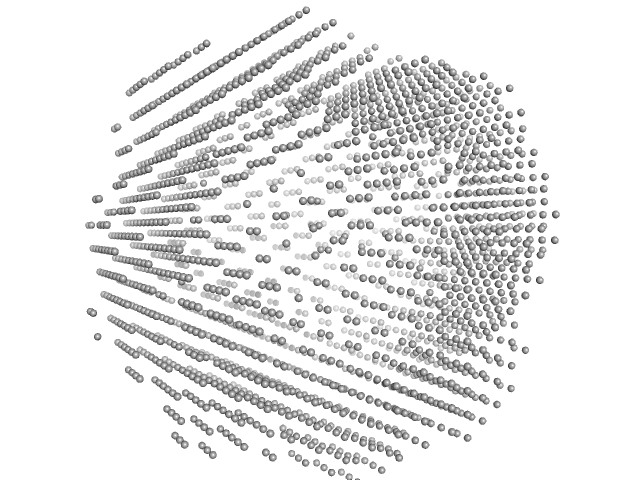
|
| Sample: |
N-terminal split fragment of a single subunit of Truncated Icosahedral Protein composed of 60-mer fusion proteins, 360 kDa synthetic construct protein
K26E mutant of C-terminal split fragment of a single subunit of Truncated Icosahedral Protein composed of 60-mer fusion proteins, 857 kDa synthetic construct protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 4 mM BaCl2, 5%(v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2023 Nov 27
|
Fusion then fission: splitting and reassembly of an artificial fusion-protein nanocage.
Chem Commun (Camb) (2024)
Ohara N, Kawakami N, Arai R, Adachi N, Ikeda A, Senda T, Miyamoto K
|
| RgGuinier |
9.4 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
3320 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Protein disulfide-isomerase monomer, 57 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris-HCL, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 4
|
Insights on the dynamic behavior of protein disulfide isomerase in the solution environment through the SAXS technique
In Silico Pharmacology 12(1) (2024)
Sanyasi C, Balakrishnan S, Chinnasamy T, Venugopalan N, Kandavelu P, Batra-Safferling R, Muthuvel S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Protein disulfide-isomerase monomer, 57 kDa Bos taurus protein
Pancreatic trypsin inhibitor monomer, 7 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris-HCL, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 4
|
Insights on the dynamic behavior of protein disulfide isomerase in the solution environment through the SAXS technique
In Silico Pharmacology 12(1) (2024)
Sanyasi C, Balakrishnan S, Chinnasamy T, Venugopalan N, Kandavelu P, Batra-Safferling R, Muthuvel S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
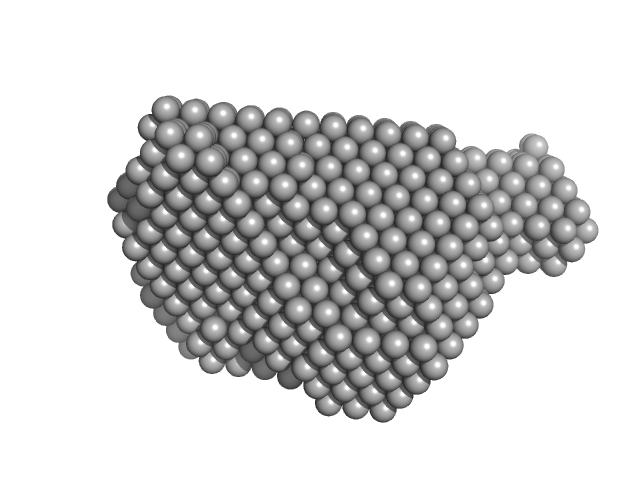
|
| Sample: |
Protein disulfide-isomerase monomer, 57 kDa Bos taurus protein
Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris-HCL, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 4
|
Insights on the dynamic behavior of protein disulfide isomerase in the solution environment through the SAXS technique
In Silico Pharmacology 12(1) (2024)
Sanyasi C, Balakrishnan S, Chinnasamy T, Venugopalan N, Kandavelu P, Batra-Safferling R, Muthuvel S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
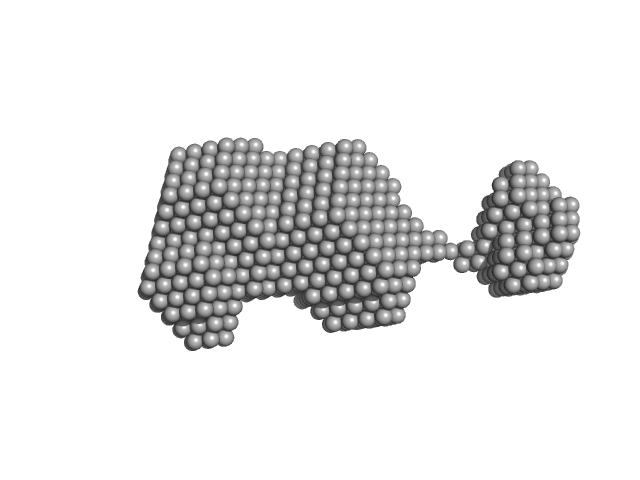
|
| Sample: |
Pancreatic trypsin inhibitor dimer, 13 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris-HCL, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 4
|
Insights on the dynamic behavior of protein disulfide isomerase in the solution environment through the SAXS technique
In Silico Pharmacology 12(1) (2024)
Sanyasi C, Balakrishnan S, Chinnasamy T, Venugopalan N, Kandavelu P, Batra-Safferling R, Muthuvel S
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
6 |
nm3 |
|
|