|
|
|
|

|
| Sample: |
Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris-HCL, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Nov 4
|
Insights on the dynamic behavior of protein disulfide isomerase in the solution environment through the SAXS technique
In Silico Pharmacology 12(1) (2024)
Sanyasi C, Balakrishnan S, Chinnasamy T, Venugopalan N, Kandavelu P, Batra-Safferling R, Muthuvel S
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GGDEF domain-containing protein dimer, 132 kDa Acidimicrobiaceae bacterium protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jan 21
|
Photocobilins integrate B(12) and bilin photochemistry for enzyme control.
Nat Commun 15(1):2740 (2024)
Zhang S, Jeffreys LN, Poddar H, Yu Y, Liu C, Patel K, Johannissen LO, Zhu L, Cliff MJ, Yan C, Schirò G, Weik M, Sakuma M, Levy CW, Leys D, Heyes DJ, Scrutton NS
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GGDEF domain-containing protein dimer, 132 kDa Acidimicrobiaceae bacterium protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jan 21
|
Photocobilins integrate B(12) and bilin photochemistry for enzyme control.
Nat Commun 15(1):2740 (2024)
Zhang S, Jeffreys LN, Poddar H, Yu Y, Liu C, Patel K, Johannissen LO, Zhu L, Cliff MJ, Yan C, Schirò G, Weik M, Sakuma M, Levy CW, Leys D, Heyes DJ, Scrutton NS
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
Replicase polyprotein 1ab (non-structural protein 16) monomer, 33 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
4.7 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|

|
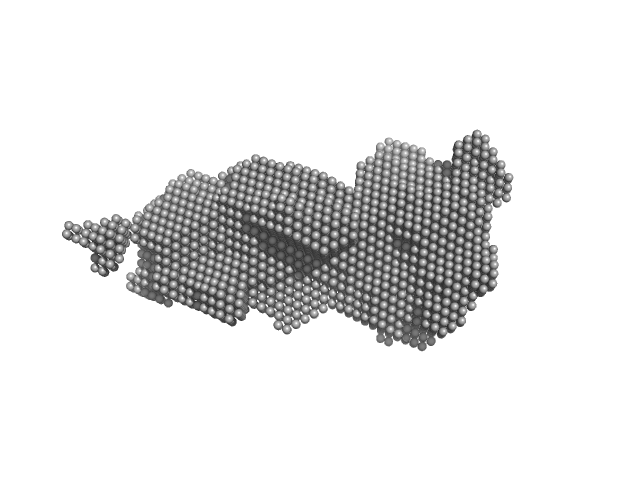
| Sample: |
Conserved protein monomer, 24 kDa Sulfolobus acidocaldarius (strain … protein
Conserved protein monomer, 44 kDa Sulfolobus acidocaldarius (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 15
|
Sequential conformational transition of ArnB, an archaeal ortholog with Sec23/Sec24 core motif
(2024)
Essen L, Korf L, Steinchen W, Watad M, Bezold F, Vogt M, Selbach L, Penner A, Tourte M, Hepp S, Albers S
|
| RgGuinier |
4.2 |
nm |
| Dmax |
19.2 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
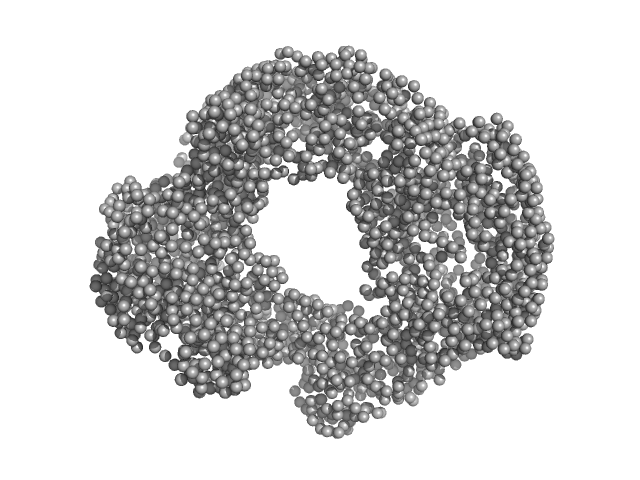
| Sample: |
Acylamino-acid-releasing enzyme (I277L, V491A) tetramer, 308 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2023 Aug 11
|
Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
FEBS Lett (2024)
Chandravanshi K, Singh R, Bhange GN, Kumar A, Yadav P, Kumar A, Makde RD
|
| RgGuinier |
5.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
424 |
nm3 |
|
|
|
|
|
|
|
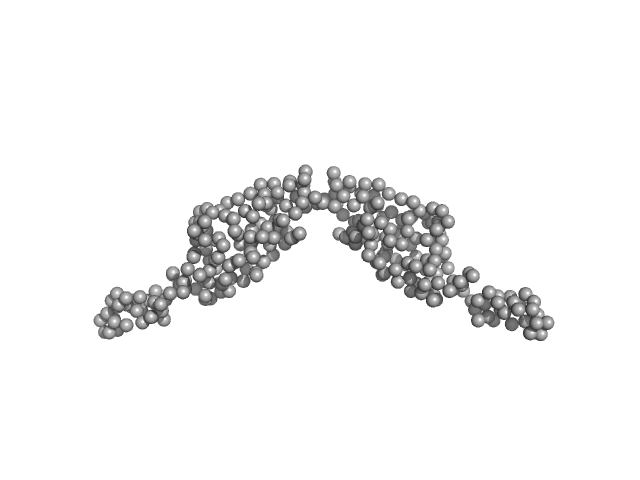
| Sample: |
Translocated intimin receptor Tir dimer, 32 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
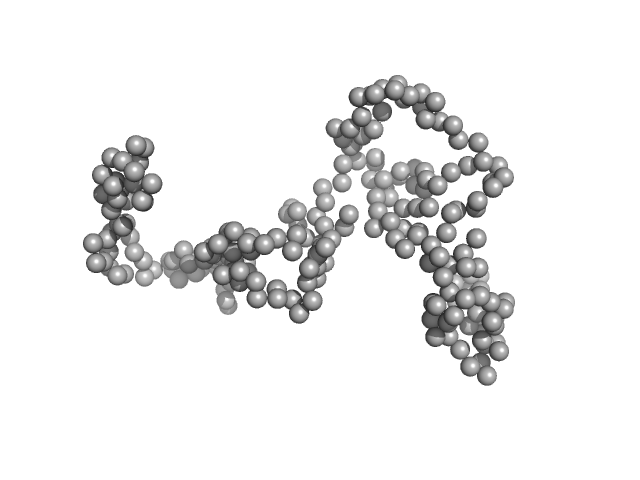
|
| Sample: |
DTMP kinase dimer, 57 kDa Brugia malayi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2018 Oct 7
|
Crystal Structure of the Brugia malayi Thymidylate Kinase-dTMP Complex and Small Angle X-ray Scattering Experiments Identifies Changes in the Dimeric Association Compared to the Human Homolog
Crystallography Reports 68(7):1150-1158 (2024)
Vishwakarma J, Sharma V, Kumar S, Ramachandran R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
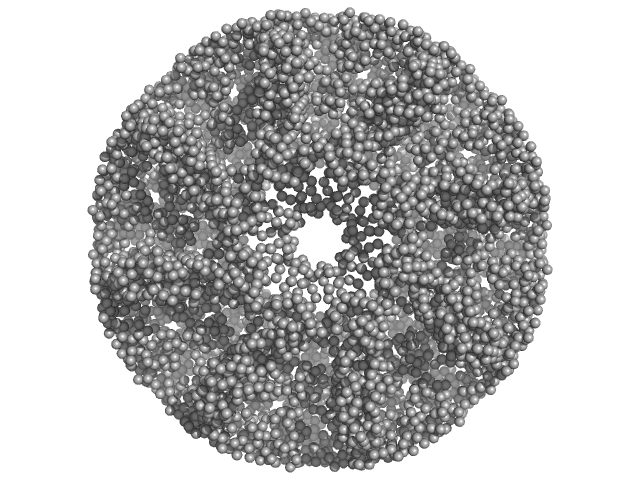
| Sample: |
Carboxypeptidase-related protein 16-mer, 903 kDa Deinococcus radiodurans (strain … protein
|
| Buffer: |
20 mM Tris-Cl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Dec 25
|
Novel oligomeric assembly of S10-carboxypeptidase from Deinococcus radiodurans
Dr. Rahul Singh
|
| RgGuinier |
7.4 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
1180 |
nm3 |
|
|
|
|
|
|
|
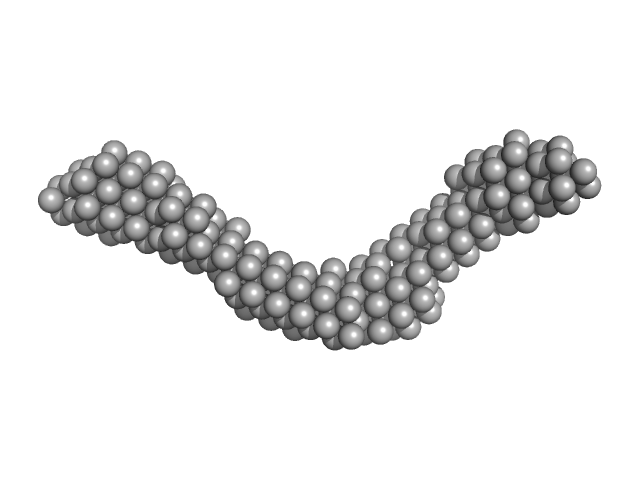
| Sample: |
Cadherin EGF LAG seven-pass G-type receptor 1 monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM KCl, 50 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Jul 1
|
CELSR1, a core planar cell polarity protein, features a weakly adhesive and flexible cadherin ectodomain.
Structure (2024)
Tamilselvan E, Sotomayor M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
85 |
nm3 |
|
|