|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein dimer, 70 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
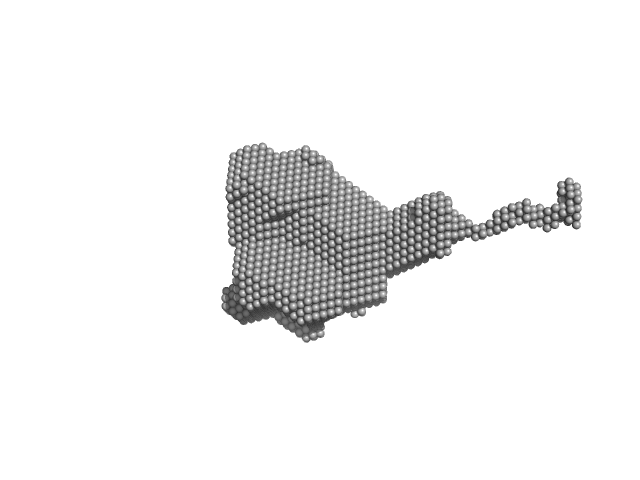
| RgGuinier |
2.8 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Orange carotenoid-binding protein dimer, 70 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein monomer, 35 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein monomer, 35 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein monomer, 35 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Orange carotenoid-binding protein monomer, 35 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
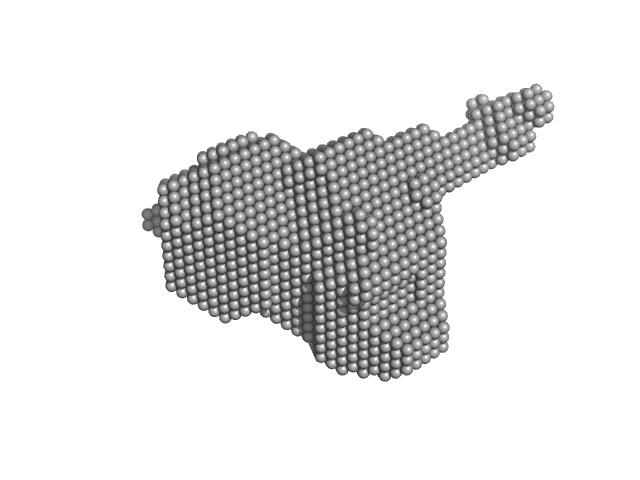
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Orange carotenoid-binding protein dimer, 70 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
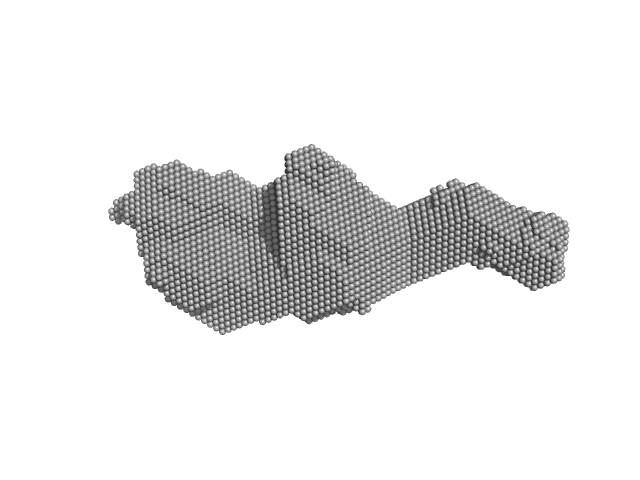
| RgGuinier |
2.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Orange carotenoid-binding protein dimer, 70 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
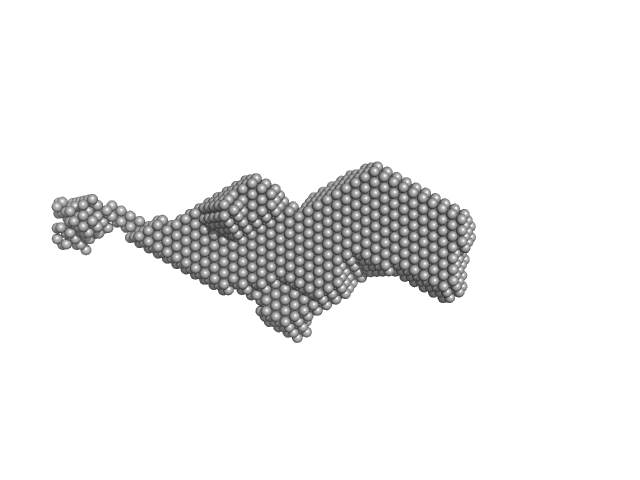
| RgGuinier |
3.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
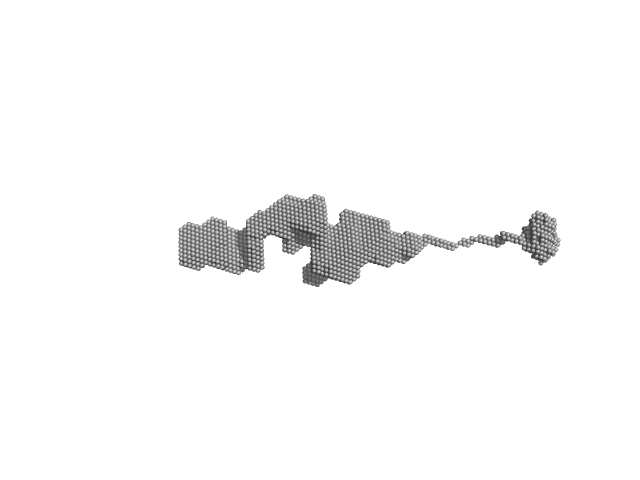
|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
4.4 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
3.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|