|
|
|
|
|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
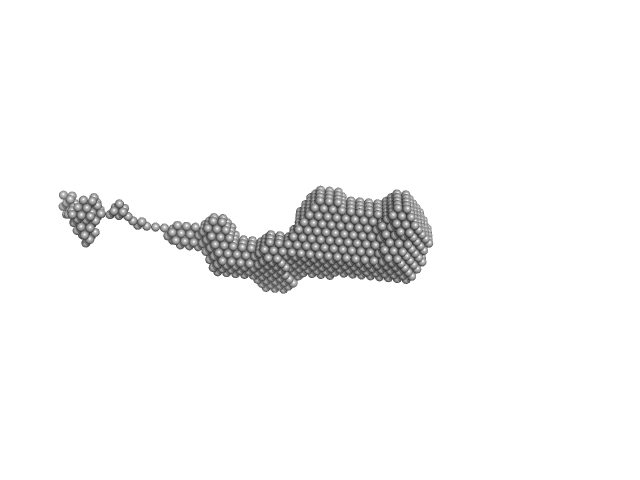
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
3.4 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
4.8 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor subfamily 1 group I member 2 monomer, 39 kDa Homo sapiens protein
Nuclear receptor subfamily 1 group I member 3 monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
25 mM Hepes, 150 mM NaCl, 5% glycerol, 5 mM DTT, pH: 7.9 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2021 Oct 8
|
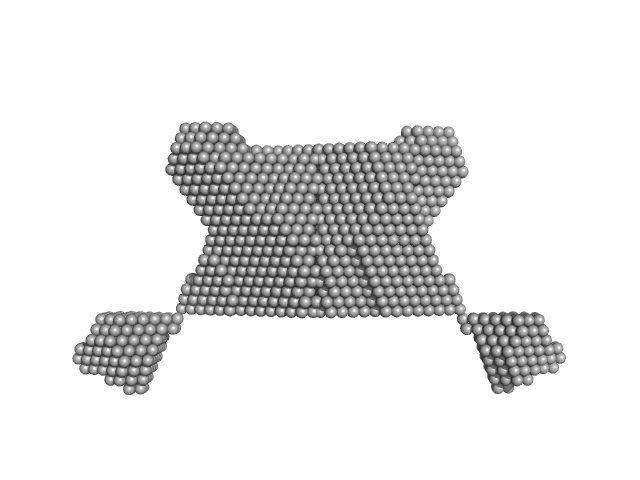
Molecular basis of crosstalk in nuclear receptors: heterodimerization between PXR and CAR and the implication in gene regulation
Shirish chodankar
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fibrillin-1 PF3 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Dec 12
|
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
5.4 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Latent-transforming growth factor beta-binding protein 1 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Dec 12
|
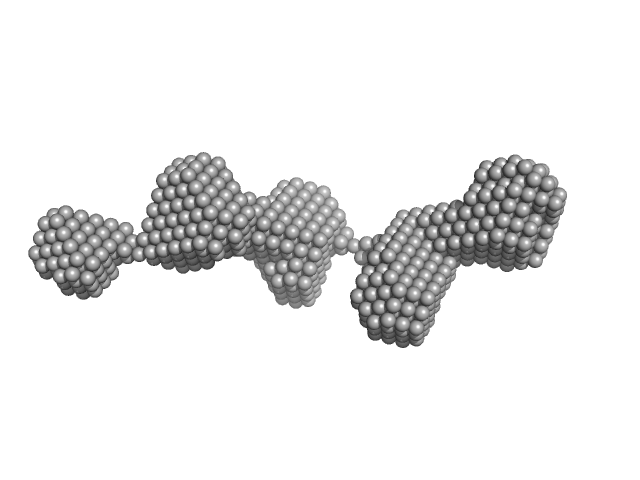
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Latent-transforming growth factor beta-binding protein 1 monomer, 44 kDa Homo sapiens protein
Fibrillin-1 PF3 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 20
|
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
7.4 |
nm |
| Dmax |
29.6 |
nm |
| VolumePorod |
759 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Kelch protein K13 (Truncated Kelch13-R539T, artemisinin-resistant mutation) hexamer, 396 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2020 Jul 25
|
Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: a SAXS data driven modeling study.
FEBS J (2022)
Goel N, Dhiman K, Kalidas N, Mukhopadhyay A, Ashish F, Bhattacharjee S
|
| RgGuinier |
6.6 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
1600 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Kelch protein K13 (Truncated Kelch13-C580Y ) hexamer, 397 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
Phosphate Buffer Saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2020 Jul 28
|
Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: a SAXS data driven modeling study.
FEBS J (2022)
Goel N, Dhiman K, Kalidas N, Mukhopadhyay A, Ashish F, Bhattacharjee S
|
| RgGuinier |
6.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
1800 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Kelch protein K13 (Truncated Kelch13-A578S) hexamer, 396 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
Phosphate Buffer Saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2020 Jul 29
|
Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: a SAXS data driven modeling study.
FEBS J (2022)
Goel N, Dhiman K, Kalidas N, Mukhopadhyay A, Ashish F, Bhattacharjee S
|
| RgGuinier |
6.1 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
1700 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Lectin nano-block WA20-H-ACG dimer, 59 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Oct 31
|
Self-Assembling Lectin Nano-Block Oligomers Enhance Binding Avidity to Glycans
International Journal of Molecular Sciences 23(2):676 (2022)
Irumagawa S, Hiemori K, Saito S, Tateno H, Arai R
|
| RgGuinier |
3.9 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|