|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
11.2 |
nm |
| Dmax |
37.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
8.6 |
nm |
| Dmax |
34.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
7.2 |
nm |
| Dmax |
18.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
5BSL3.2 monomer, 15 kDa Hepatitis C virus … RNA
|
| Buffer: |
10 mM Tris-HCl, 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2019 Jun 2
|
The low-resolution structural models of hepatitis C virus RNA subdomain 5BSL3.2 and its distal complex with domain 3'X point to conserved regulatory mechanisms within the Flaviviridae family.
Nucleic Acids Res (2022)
Castillo-Martínez J, Fan L, Szewczyk MP, Wang YX, Gallego J
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
5BSL3.2 monomer, 15 kDa Hepatitis C virus … RNA
|
| Buffer: |
10 mM Tris-HCl, 6 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2019 Jun 2
|
The low-resolution structural models of hepatitis C virus RNA subdomain 5BSL3.2 and its distal complex with domain 3'X point to conserved regulatory mechanisms within the Flaviviridae family.
Nucleic Acids Res (2022)
Castillo-Martínez J, Fan L, Szewczyk MP, Wang YX, Gallego J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
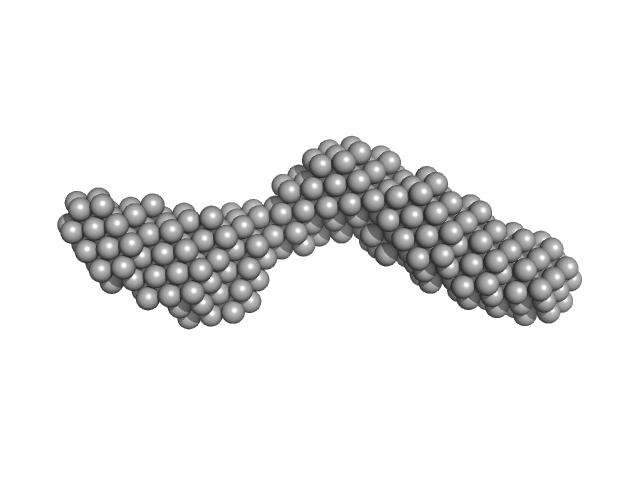
|
| Sample: |
Domain 3'X mutant monomer, 32 kDa Hepatitis C virus … RNA
|
| Buffer: |
10 mM Tris-HCl, 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2019 Jun 2
|
The low-resolution structural models of hepatitis C virus RNA subdomain 5BSL3.2 and its distal complex with domain 3'X point to conserved regulatory mechanisms within the Flaviviridae family.
Nucleic Acids Res (2022)
Castillo-Martínez J, Fan L, Szewczyk MP, Wang YX, Gallego J
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Domain 3'X mutant monomer, 32 kDa Hepatitis C virus … RNA
|
| Buffer: |
10 mM Tris-HCl, 6 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jun 2
|
The low-resolution structural models of hepatitis C virus RNA subdomain 5BSL3.2 and its distal complex with domain 3'X point to conserved regulatory mechanisms within the Flaviviridae family.
Nucleic Acids Res (2022)
Castillo-Martínez J, Fan L, Szewczyk MP, Wang YX, Gallego J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Domain 3'X mutant - 5BSL3.2 complex monomer, 42 kDa Hepatitis C virus … RNA
|
| Buffer: |
10 mM Tris-HCl, 6 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2019 Jun 2
|
The low-resolution structural models of hepatitis C virus RNA subdomain 5BSL3.2 and its distal complex with domain 3'X point to conserved regulatory mechanisms within the Flaviviridae family.
Nucleic Acids Res (2022)
Castillo-Martínez J, Fan L, Szewczyk MP, Wang YX, Gallego J
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein monomer, 35 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein dimer, 70 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
77 |
nm3 |
|
|