|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDG72_fit1_model1.png)
|
| Sample: |
1,2-dimyristoyl-sn-glycero-3-phosphocholine monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 9
|
Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data
Journal of Applied Crystallography 54(1) (2021)
Konarev P, Gruzinov A, Mertens H, Svergun D
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDG82_fit1_model1.png)
|
| Sample: |
1,2-dimyristoyl-sn-glycero-3-phosphocholine monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 9
|
Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data
Journal of Applied Crystallography 54(1) (2021)
Konarev P, Gruzinov A, Mertens H, Svergun D
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDG92_fit1_model1.png)
|
| Sample: |
1,2-dipalmitoyl-sn-glycero-3-phosphocholine monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 9
|
Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data
Journal of Applied Crystallography 54(1) (2021)
Konarev P, Gruzinov A, Mertens H, Svergun D
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGA2_fit1_model1.png)
|
| Sample: |
1,2-dipalmitoyl-sn-glycero-3-phosphocholine monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 9
|
Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data
Journal of Applied Crystallography 54(1) (2021)
Konarev P, Gruzinov A, Mertens H, Svergun D
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGB2_fit1_model1.png)
|
| Sample: |
1,2-dipalmitoyl-sn-glycero-3-phosphocholine monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 9
|
Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data
Journal of Applied Crystallography 54(1) (2021)
Konarev P, Gruzinov A, Mertens H, Svergun D
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDGC2_fit1_model1.png)
|
| Sample: |
1,2-dipalmitoyl-sn-glycero-3-phosphocholine monomer, 1 kDa
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 9
|
Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data
Journal of Applied Crystallography 54(1) (2021)
Konarev P, Gruzinov A, Mertens H, Svergun D
|
|
|
|
|
|
|
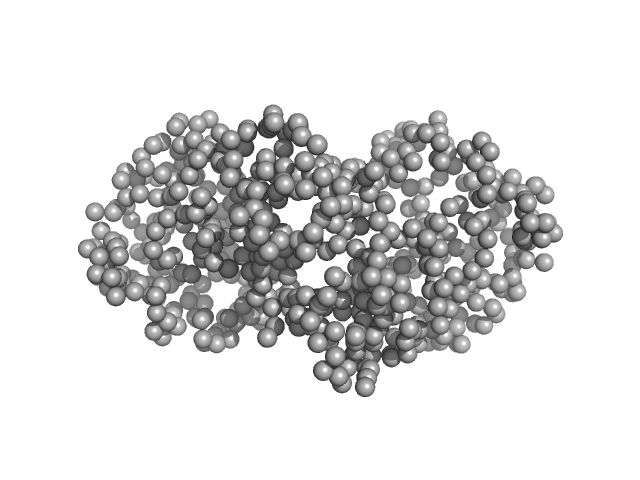
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆295-358 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 21
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
101.6 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
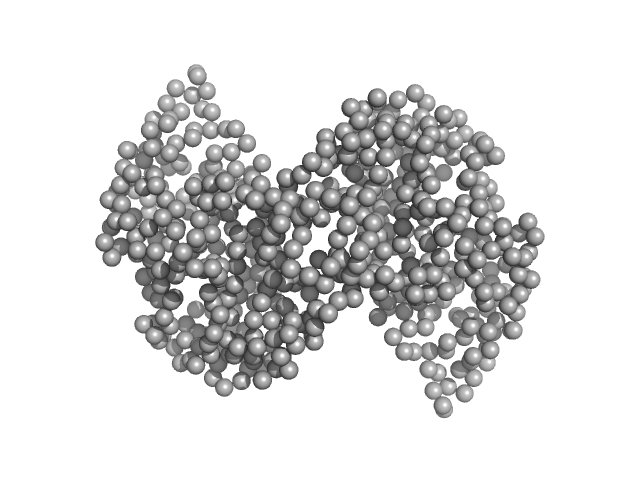
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆295-358 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆295-358 monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 21
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
92.3 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
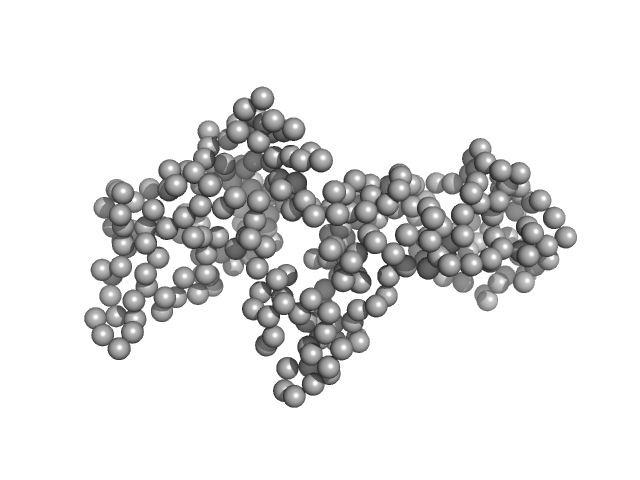
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆295-358 monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|