|
|
|
|
|
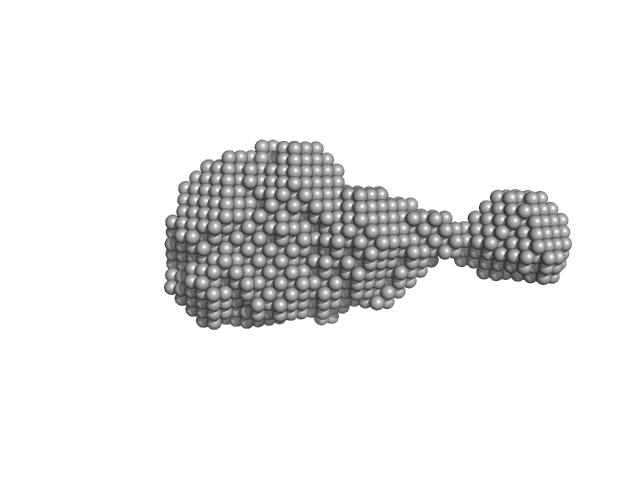
| Sample: |
4-O-methyl-glucuronoyl methylesterase (Glucuronoyl esterase) monomer, 51 kDa Cerrena unicolor protein
|
| Buffer: |
20 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 10
|
The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun 11(1):1026 (2020)
Ernst HA, Mosbech C, Langkilde AE, Westh P, Meyer AS, Agger JW, Larsen S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human Telomerase Reverse Transcriptase Core Promoter G-rich Region monomer, 22 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Oct 5
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Antiparallel Hairpin monomer, 21 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 3
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Optimized Parallel monomer, 18 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 3
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HTERT core promoter PQS2-PQS3 region monomer, 14 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 3
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Antiparallel Hairpin PQS2-PQS3 segment monomer, 14 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 3
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 14 kDa Gallus gallus protein
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 5
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
30.2 |
nm |
| Dmax |
120.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; radius 5.6 nm (AFM based) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 8
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
29.4 |
nm |
| Dmax |
80.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Calcium-activated chloride channel regulator 1 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jun 6
|
Structural and Biophysical Analysis of the CLCA1 VWA Domain Suggests Mode of TMEM16A Engagement.
Cell Rep 30(4):1141-1151.e3 (2020)
Berry KN, Brett TJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
Calcium-activated chloride channel regulator 1 monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 6
|
Structural and Biophysical Analysis of the CLCA1 VWA Domain Suggests Mode of TMEM16A Engagement.
Cell Rep 30(4):1141-1151.e3 (2020)
Berry KN, Brett TJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.4 |
nm |
|
|