|
|
|
|
|
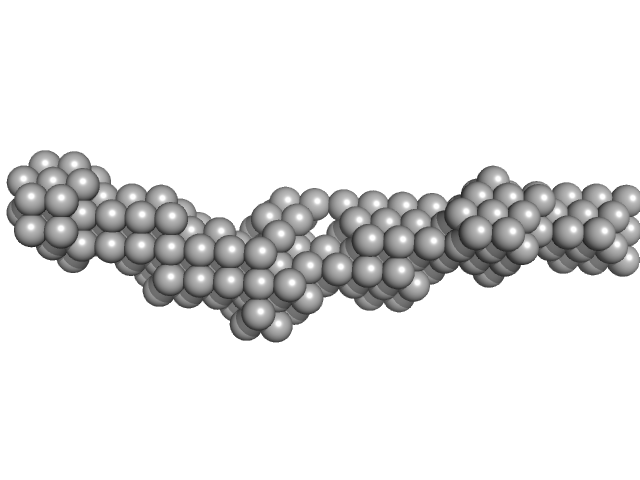
| Sample: |
Meiotic localizer of BRCA2 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Probable exodeoxyribonuclease III protein XthA monomer, 32 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl 500 mM NaCl 5mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 12
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
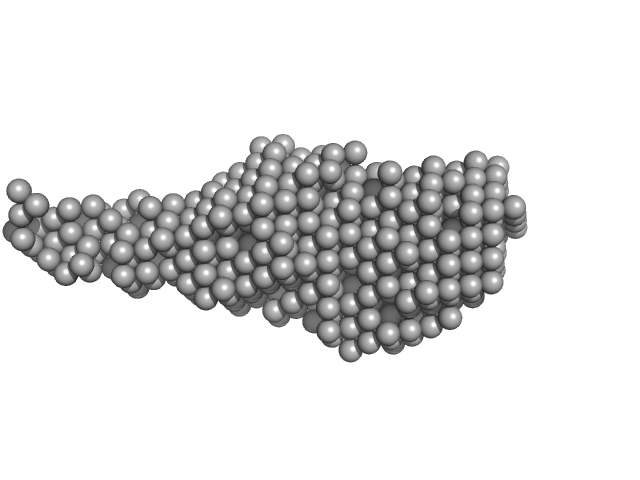
| Sample: |
Probable exodeoxyribonuclease III protein XthA monomer, 33 kDa Mycobacterium tuberculosis protein
M. tb. LigA BRCT domain monomer, 15 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl 500 mM NaCl 5mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 9
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 13
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
870 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Nicked DNA dimer, 16 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 13
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
|
|
|
|
|
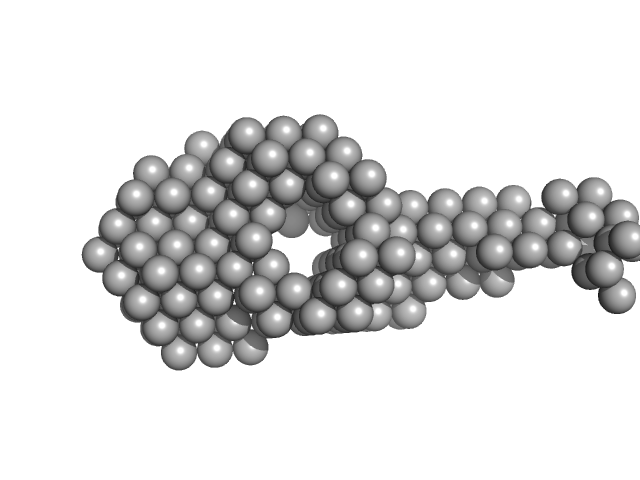
| Sample: |
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Probable exodeoxyribonuclease III protein XthA monomer, 33 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 13
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.9 |
nm |
| VolumePorod |
662 |
nm3 |
|
|
|
|
|
|
|
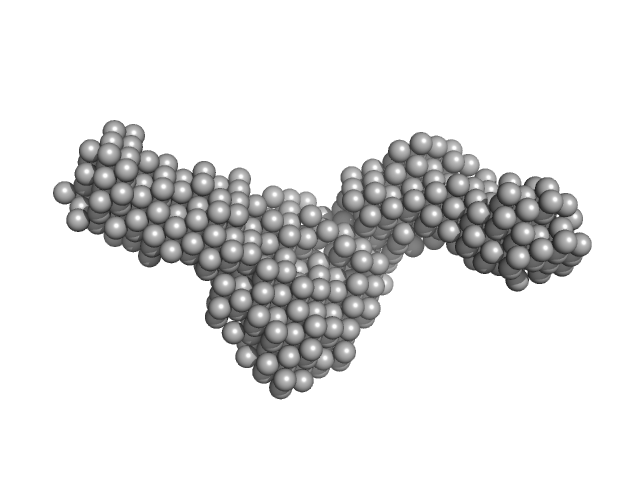
| Sample: |
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Probable exodeoxyribonuclease III protein XthA monomer, 33 kDa Mycobacterium tuberculosis protein
Nicked DNA monomer, 8 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 13
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
TRAF-interacting protein with FHA domain-containing protein A dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 100 mM arginine, 5 % glycerol, 10 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Kumamoto University on 2014 Oct 16
|
Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep 10(1):5152 (2020)
Nakamura T, Hashikawa C, Okabe K, Yokote Y, Chirifu M, Toma-Fukai S, Nakamura N, Matsuo M, Kamikariya M, Okamoto Y, Gohda J, Akiyama T, Semba K, Ikemizu S, Otsuka M, Inoue JI, Yamagata Y
|
| RgGuinier |
3.1 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
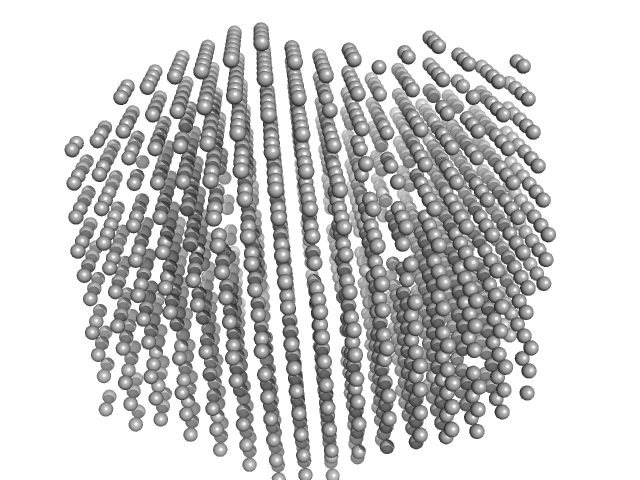
| Sample: |
2,4-dichlorophenol 6-monooxygenase hexamer, 399 kDa Streptomyces sp. SCSIO … protein
Flavin adenine dinucleotide hexamer, 5 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5 mM DTT, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Oct 22
|
Structural analyses of the group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J Biol Chem (2020)
Manenda MS, Picard MÈ, Zhang L, Cyr N, Zhu X, Barma J, Pascal JM, Couture M, Zhang C, Shi R
|
| RgGuinier |
4.8 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
624 |
nm3 |
|
|
|
|
|
|
|
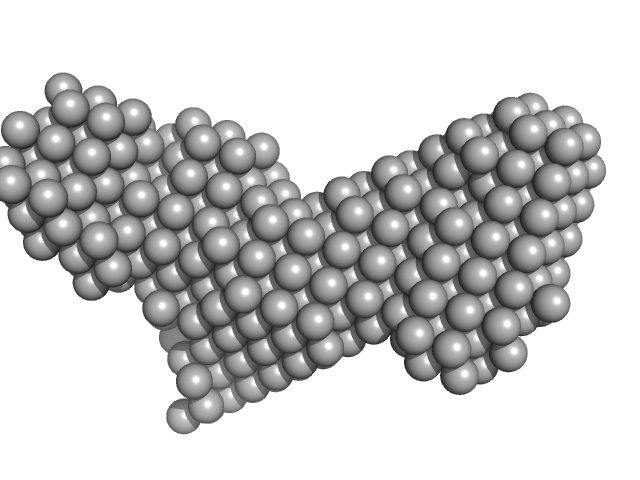
| Sample: |
Histone deacetylase 1 monomer, 55 kDa Homo sapiens protein
Lysine-specific histone demethylase 1A monomer, 93 kDa Homo sapiens protein
REST corepressor 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris/Cl, 50 mM potassium acetate and 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Jan 23
|
Mechanism of Crosstalk between the LSD1 Demethylase and HDAC1 Deacetylase in the CoREST Complex.
Cell Rep 30(8):2699-2711.e8 (2020)
Song Y, Dagil L, Fairall L, Robertson N, Wu M, Ragan TJ, Savva CG, Saleh A, Morone N, Kunze MBA, Jamieson AG, Cole PA, Hansen DF, Schwabe JWR
|
| RgGuinier |
6.0 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
437 |
nm3 |
|
|