|
|
|
|

|
| Sample: |
Xrn1 resistance RNA-1 from West Nile virus monomer, 25 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xrn1 resistance RNA-1 from Murray Valley Encephalitis monomer, 26 kDa Murray Valley Encephalitis RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
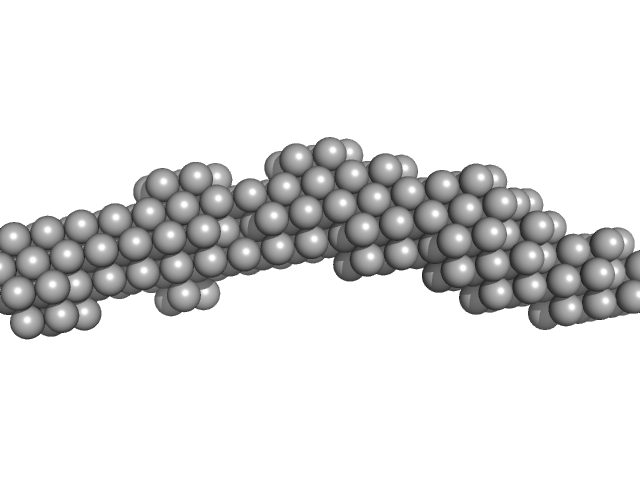
| Sample: |
3'SL from Dengue virus 2 monomer, 31 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Feb 24
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
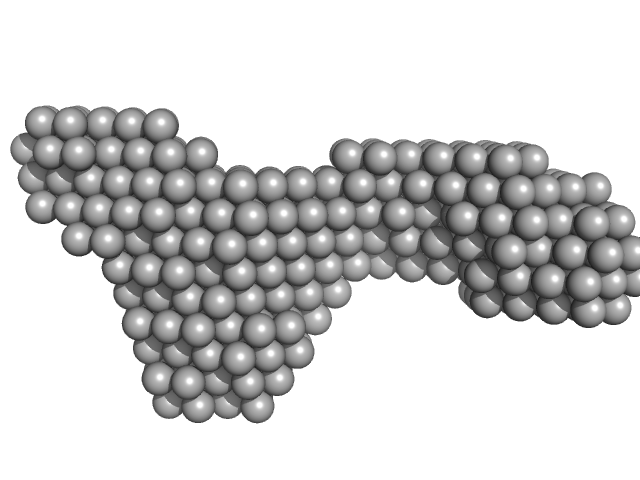
| Sample: |
Xrn1 resistance RNA1-2 from Dengue virus 2 monomer, 46 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xrn1 resistance RNA2 from Zika virus monomer, 22 kDa Zika virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2017 Sep 11
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xrn1 resistance RNA1-2 from West Nile virus monomer, 75 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Apr 2
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
235 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SL3 from West Nile virus monomer, 23 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2017 Jun 27
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xrn1 resistance RNA2 from Dengue virus 2 monomer, 21 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Mar 16
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DB12 from Dengue virus 2 monomer, 56 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Feb 24
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xrn1 resistance RNA2 from West Nile virus monomer, 23 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2017 Apr 7
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
30 |
nm3 |
|
|