|
|
|
|

|
| Sample: |
Xrn1 resistance RNA2 from Murray Valley Encephalitis monomer, 22 kDa Murray Valley Encephalitis RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 14
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DB12 from Zika virus monomer, 47 kDa Zika virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Apr 2
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
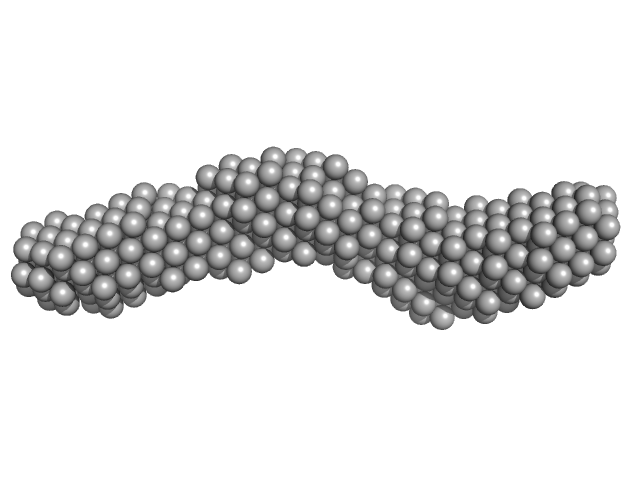
| Sample: |
DB12 from West Nile virus monomer, 59 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jan 19
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
3'SL from Zika virus monomer, 32 kDa Zika virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
3'SL from West Nile virus monomer, 31 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2017 Apr 7
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
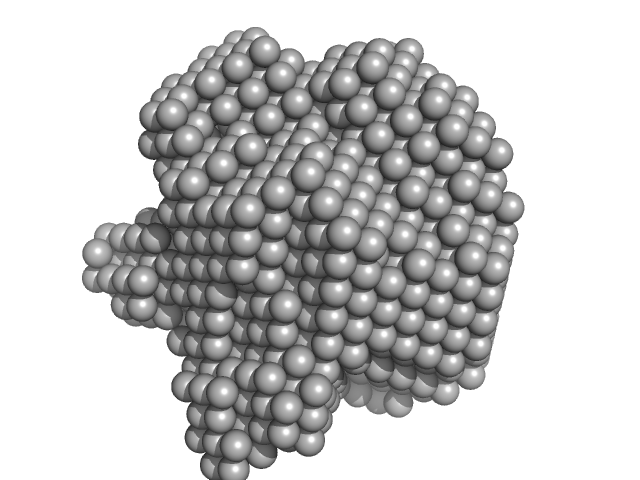
| Sample: |
Methylxanthine N1-demethylase NdmA trimer, 127 kDa Pseudomonas putida protein
Methylxanthine N3-demethylase NdmB trimer, 129 kDa Pseudomonas putida protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 2 mM TCEP 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2018 Jul 27
|
Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J Mol Biol 431(19):3647-3661 (2019)
Kim JH, Kim BH, Brooks S, Kang SY, Summers RM, Song HK
|
| RgGuinier |
4.5 |
nm |
| Dmax |
12.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
Methylxanthine N1-demethylase NdmA trimer, 207 kDa Pseudomonas putida protein
Methylxanthine N3-demethylase NdmB trimer, 129 kDa Pseudomonas putida protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 2 mM TCEP 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2018 Jul 27
|
Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J Mol Biol 431(19):3647-3661 (2019)
Kim JH, Kim BH, Brooks S, Kang SY, Summers RM, Song HK
|
| RgGuinier |
5.4 |
nm |
| Dmax |
13.8 |
nm |
|
|
|
|
|
|

|
| Sample: |
Methylxanthine N1-demethylase NdmA hexamer, 254 kDa Pseudomonas putida protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 May 21
|
Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J Mol Biol 431(19):3647-3661 (2019)
Kim JH, Kim BH, Brooks S, Kang SY, Summers RM, Song HK
|
| RgGuinier |
4.2 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Methylxanthine N3-demethylase NdmB hexamer, 258 kDa Pseudomonas putida protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 May 21
|
Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J Mol Biol 431(19):3647-3661 (2019)
Kim JH, Kim BH, Brooks S, Kang SY, Summers RM, Song HK
|
| RgGuinier |
4.3 |
nm |
| Dmax |
12.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
Methylxanthine N1-demethylase NdmA trimer, 127 kDa Pseudomonas putida protein
Methylxanthine N3-demethylase NdmB trimer, 129 kDa Pseudomonas putida protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 May 21
|
Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex.
J Mol Biol 431(19):3647-3661 (2019)
Kim JH, Kim BH, Brooks S, Kang SY, Summers RM, Song HK
|
| RgGuinier |
4.2 |
nm |
| Dmax |
10.9 |
nm |
|
|