|
|
|
|
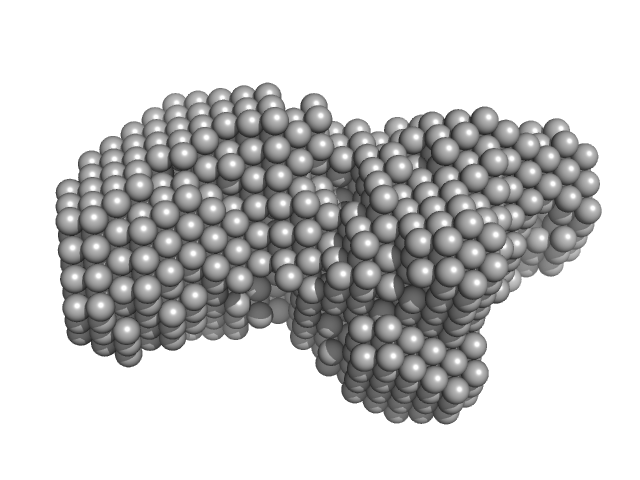
|
| Sample: |
Enhanced disease susceptibility monomer, 72 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM NaCl, 50 mM HEPES, 1% glyercol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 May 19
|
Arabidopsis immunity regulator EDS1 in a PAD4/SAG101-unbound form is a monomer with an inherently inactive conformation.
J Struct Biol :107390 (2019)
Voss M, Toelzer C, Bhandari DD, Parker JE, Niefind K
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
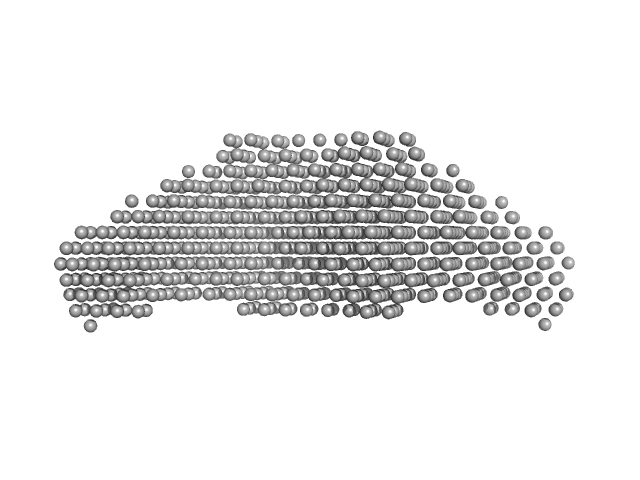
|
| Sample: |
STI1-like protein dimer, 136 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2016 Aug 3
|
Structural studies of the Hsp70/Hsp90 organizing protein of Plasmodium falciparum and its modulation of Hsp70 and Hsp90 ATPase activities.
Biochim Biophys Acta Proteins Proteom :140282 (2019)
Silva NSM, Bertolino-Reis DE, Dores-Silva PR, Anneta FB, Seraphim TV, Barbosa LRS, Borges JC
|
| RgGuinier |
6.3 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
557 |
nm3 |
|
|
|
|
|
|
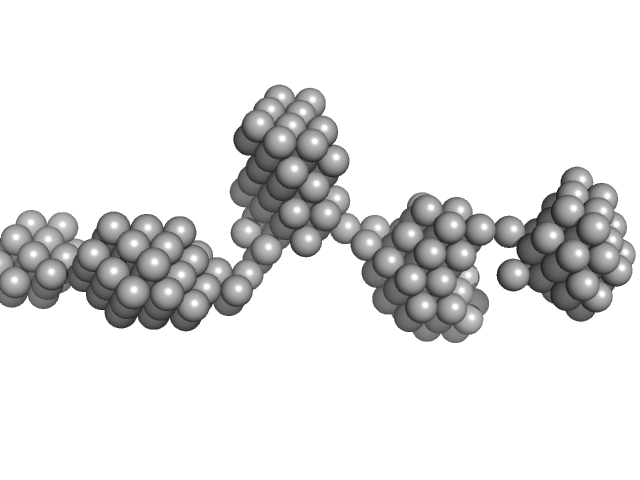
|
| Sample: |
Aryl hydrocarbon receptor nuclear translocator-like protein 1 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|
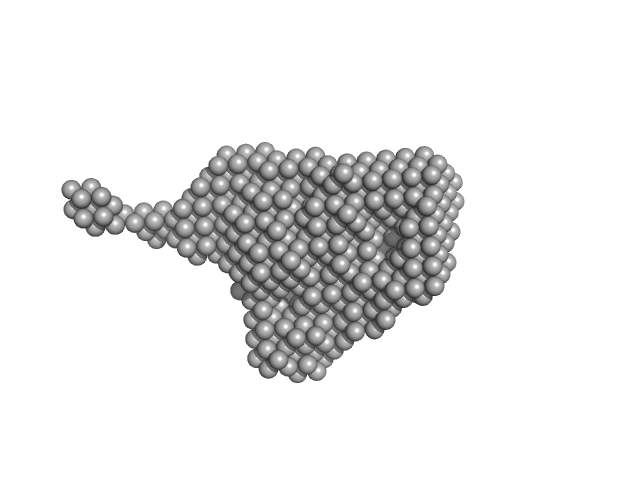
|
| Sample: |
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP), mutation L664C monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
1.9 |
nm |
| Dmax |
8.1 |
nm |
|
|
|
|
|
|

|
| Sample: |
Aryl hydrocarbon receptor nuclear translocator-like protein 1 monomer, 10 kDa Mus musculus protein
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP), mutation L664C monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Brain and muscle ARNT-like 1 (G517-L625) including transactivation domain (TAD) monomer, 11 kDa Mus musculus protein
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP) monomer, 10 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP) monomer, 10 kDa Mus musculus protein
Brain and muscle ARNT-like 1 (G490-L625) including transactivation domain (TAD) monomer, 14 kDa Mus musculus protein
|
| Buffer: |
25 mM Hepes, 150 NaCl, 1 mM DTT, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
17.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Kinase-inducible domain interacting (KIX) domain of CREB-binding protein (CBP), mutation L664C monomer, 10 kDa Mus musculus protein
1-{4-[4-chloro-3-(trifluoromethyl)phenyl]-4-hydroxypiperidin-1-yl}-3-sulfanylpropan-1-one monomer, 0 kDa
|
| Buffer: |
25 mM Hepes, 150 NaCl, 5% Glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 27
|
Structural and mechanistic insights into the interaction of the circadian transcription factor BMAL1 with the KIX domain of the CREB-binding protein.
J Biol Chem (2019)
Garg A, Orru R, Ye W, Distler U, Chojnacki JE, Köhn M, Tenzer S, Sönnichsen C, Wolf E
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.4 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xrn1 resistance RNA1 from Dengue virus 2 monomer, 21 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xrn1 resistance RNA1-2 from Dengue virus 2 monomer, 46 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Apr 2
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
67 |
nm3 |
|
|