|
|
|
|

|
| Sample: |
Histone H3.3 monomer, 4 kDa Homo sa protein
DNA (cytosine-5)-methyltransferase 3B Pro-Trp-Trp-Pro (PWWP) domain monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2024 Sep 17
|
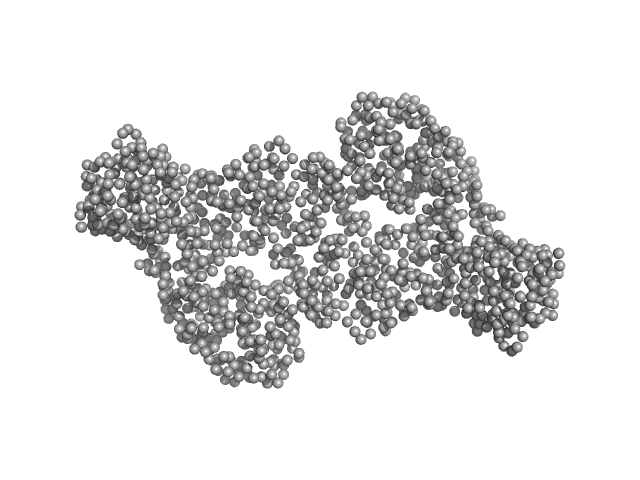
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
DNA methyltransferase 3 beta (413-853) dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Nov 2
|
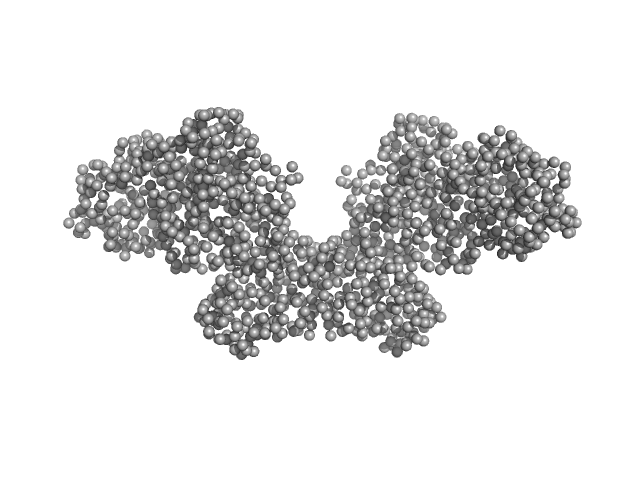
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
191 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histone H3.3 dimer, 8 kDa Homo sapiens protein
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
DNA methyltransferase 3 beta (413-853) dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Nov 2
|
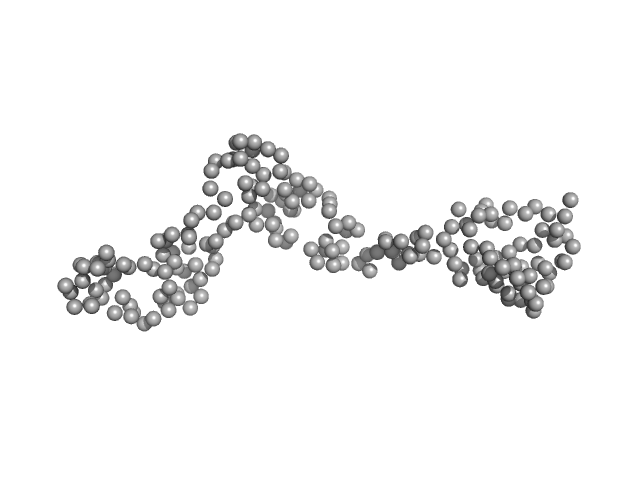
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
4.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 21 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 42 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
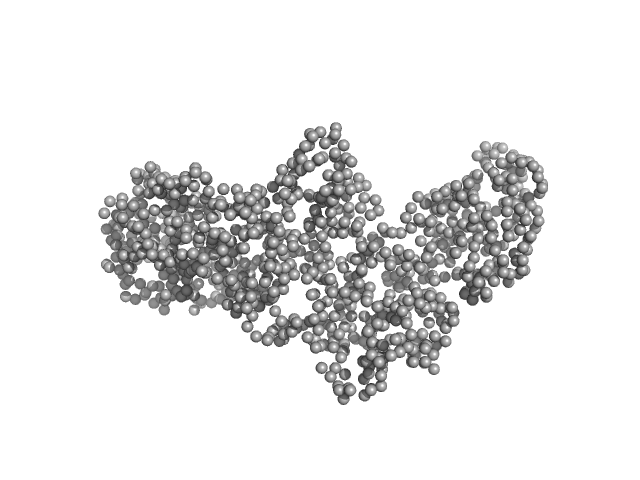
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
25-mer dsRNA monomer, 16 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.9 |
nm |
| VolumePorod |
453 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
25-mer dsRNA monomer, 16 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
431 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
36-mer dsRNA monomer, 23 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
587 |
nm3 |
|
|