|
|
|
|

|
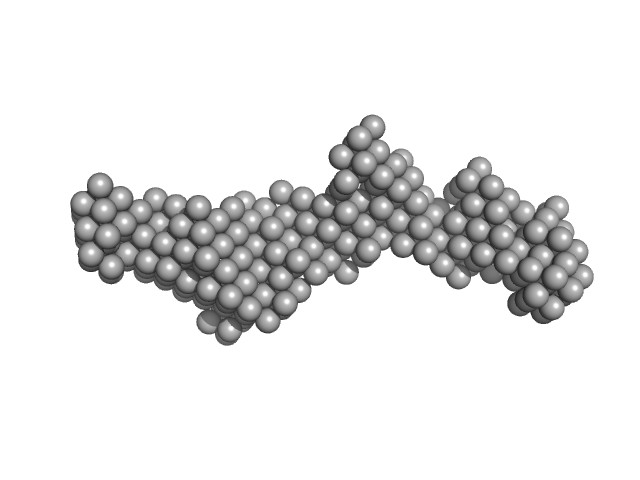
| Sample: |
EB120 RNA monomer monomer, 39 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
BC120 G25C RNA monomer, 39 kDa Homo sapiens RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
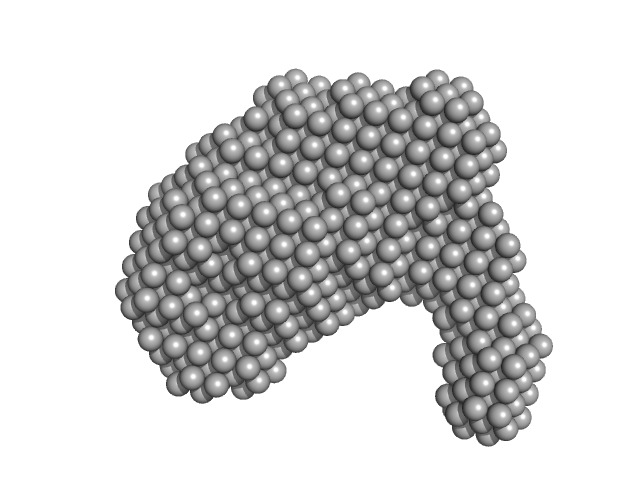
| Sample: |
ESX-5 secretion-associated protein EspG5 monomer, 32 kDa Mycobacterium tuberculosis (strain … protein
ESX-5 secretion system protein EccA5 monomer, 31 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2025 Jan 28
|
Small angle x-ray scattering envelope of EccA5 N-terminal domain and EspG5 of Type 7 secretion pathway of Mycobacterium tuberculosis
Rajlakshmi K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (L353P) dimer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 7
|
The CRISPR-Cas9 knockout DDC SH-SY5Y in vitro model for AADC deficiency provides insight into the pathogenicity of R347Q and L353P variants: a cross-sectional structural and functional analysis.
FEBS J (2025)
Carmona-Carmona CA, Bisello G, Franchini R, Lunardi G, Galavotti R, Perduca M, Ribeiro RP, Belviso BD, Giorgetti A, Caliandro R, Lievens PM, Bertoldi M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (R347Q) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 µM pyridoxal 5'-phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Jul 7
|
The CRISPR-Cas9 knockout DDC SH-SY5Y in vitro model for AADC deficiency provides insight into the pathogenicity of R347Q and L353P variants: a cross-sectional structural and functional analysis.
FEBS J (2025)
Carmona-Carmona CA, Bisello G, Franchini R, Lunardi G, Galavotti R, Perduca M, Ribeiro RP, Belviso BD, Giorgetti A, Caliandro R, Lievens PM, Bertoldi M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
|
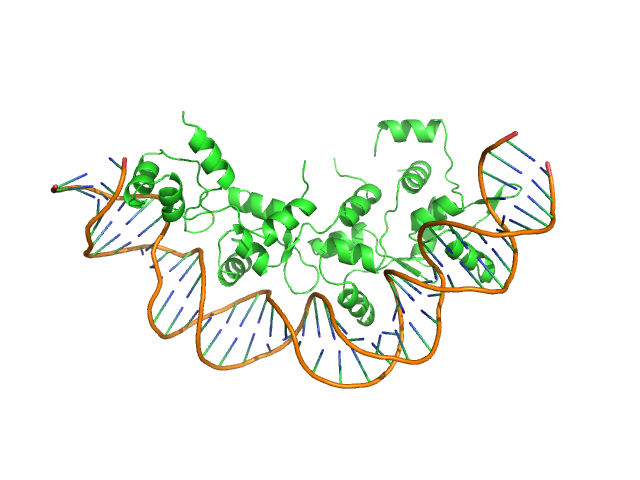
| Sample: |
Recombination directionality factor RdfS tetramer, 52 kDa Mesorhizobium japonicum R7A protein
AttP_8 40-mer DNA dimer, 25 kDa DNA
|
| Buffer: |
150 mM Tris-HCl, 300 mM NaCl, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 19
|
Structural basis for control of integrative and conjugative element excision and transfer by the oligomeric winged helix–turn–helix protein RdfS
Nucleic Acids Research 53(6) (2025)
Verdonk C, Agostino M, Eto K, Hall D, Bond C, Ramsay J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDVV6_fit1_model1.png)
|
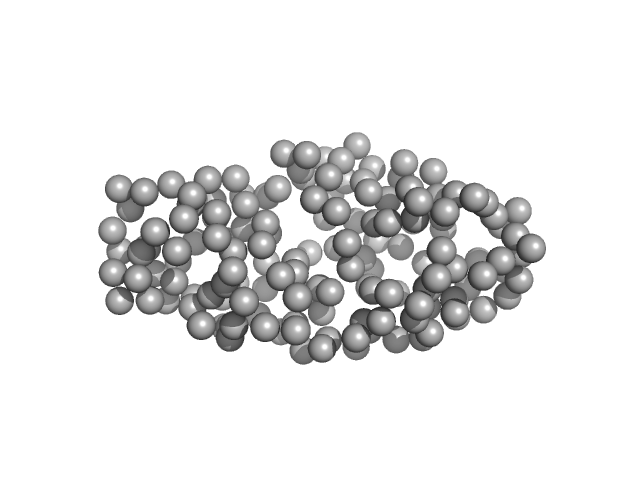
| Sample: |
Drebrin monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
17 mM NaH2PO4, 3 mM Na2HPO4, 50 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jul 7
|
Dynamic Interchange of Local Residue-Residue Interactions in the Largely Extended Single Alpha-Helix in Drebrin
Biochemical Journal (2025)
Varga S, Péterfia B, Dudola D, Farkas V, Jeffries C, Permi P, Gáspári Z
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA (cytosine-5)-methyltransferase 3B Pro-Trp-Trp-Pro (PWWP) domain monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2024 Apr 20
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
20464 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
DNA methyltransferase 3-like (178-379) dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
271263 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Histone H3.3 dimer, 8 kDa Homo sapiens protein
DNA methyltransferase 3 beta (215-853) dimer, 145 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 1 mM TCEP, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 16
|
Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation.
Sci Adv 11(13):eadu8116 (2025)
Cho CC, Huang HH, Jiang BC, Yang WZ, Chen YN, Yuan HS
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
275 |
nm3 |
|
|