|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
36-mer dsRNA monomer, 23 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.1 |
nm |
| VolumePorod |
626 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.1 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
797 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.7 |
nm |
| Dmax |
26.5 |
nm |
| VolumePorod |
1075 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.8 |
nm |
| Dmax |
26.1 |
nm |
| VolumePorod |
1050 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
MOE PS gapmers 3-8-3 monomer, 4 kDa DNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
MOE PS gapmers 3-8-3 monomer, 4 kDa DNA
PSCK9, 24mer ASO binding site monomer, 8 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Oct 21
|
Application of enhanced biophysical strategies to determine ASO/target RNA binding affinity and kinetics
Michael Lerche
|
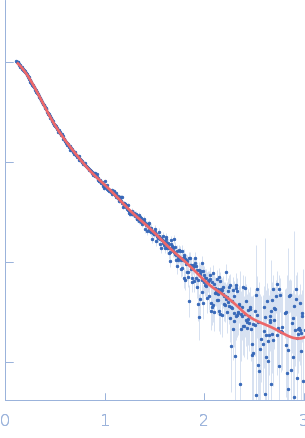
| RgGuinier |
2.1 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endophilin-B1 dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
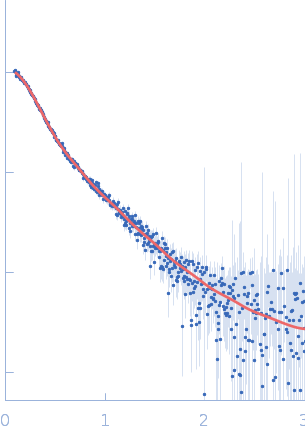
| RgGuinier |
5.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endophilin-B1 dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
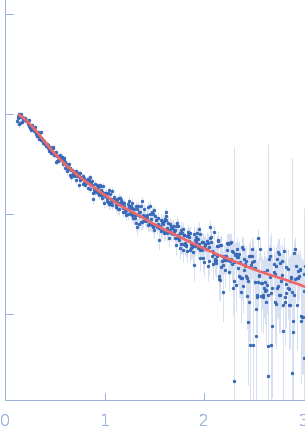
| RgGuinier |
5.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endophilin-B1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
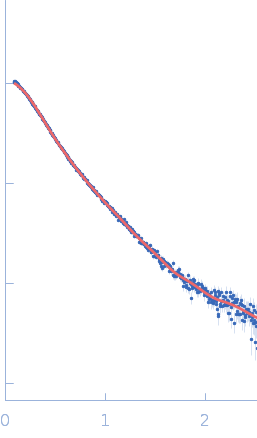
| RgGuinier |
3.7 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endophilin-B1 (Δ307-360) dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
149 |
nm3 |
|
|