|
|
|
|

|
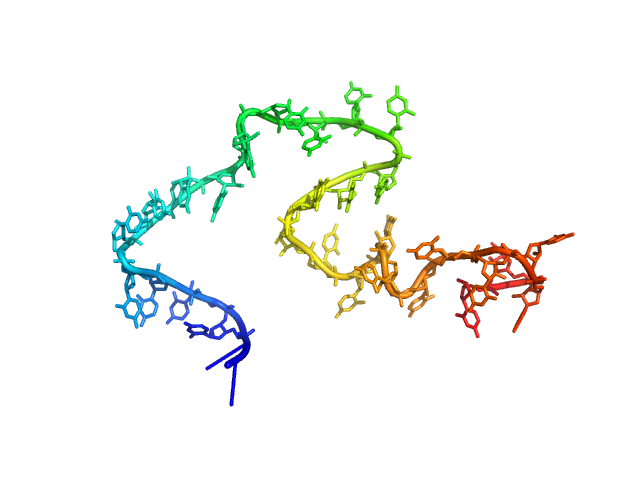
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 100 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
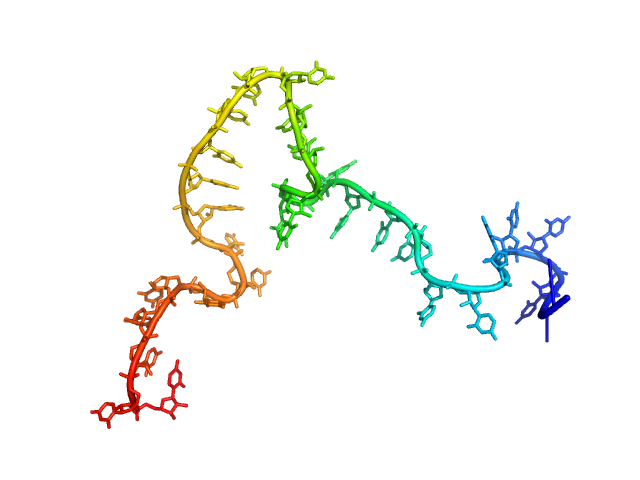
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 200 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
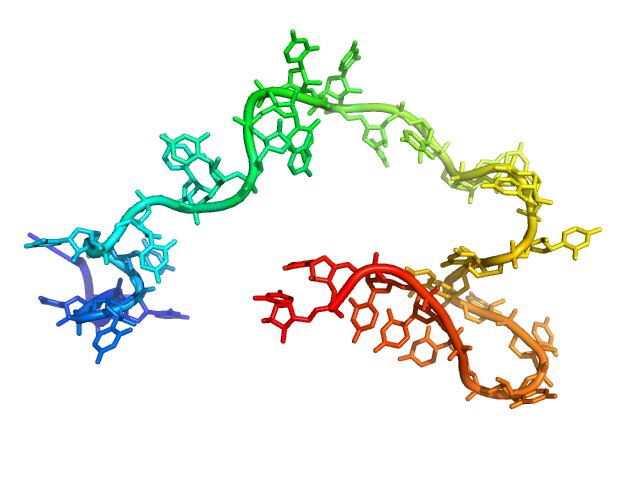
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 1 mM MgCl2, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
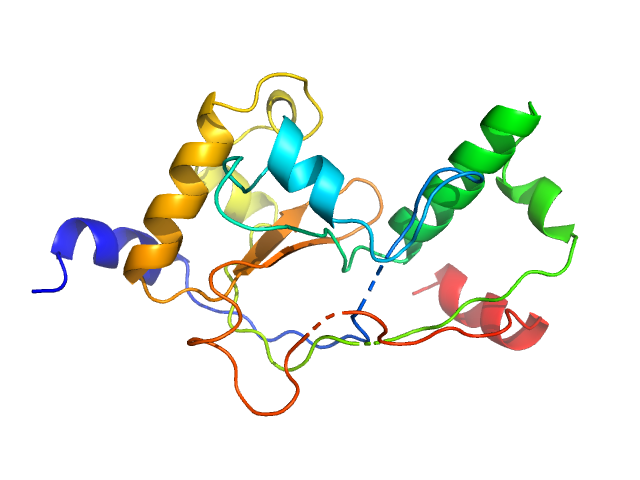
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 2 mM MgCl2, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mycobacterial cidal toxin monomer, 20 kDa Mycobacterium tuberculosis H37Rv protein
|
| Buffer: |
30 mM Tris-HCl, 200 mM NaCl, 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 2
|
MbsTA
Diana Freire
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
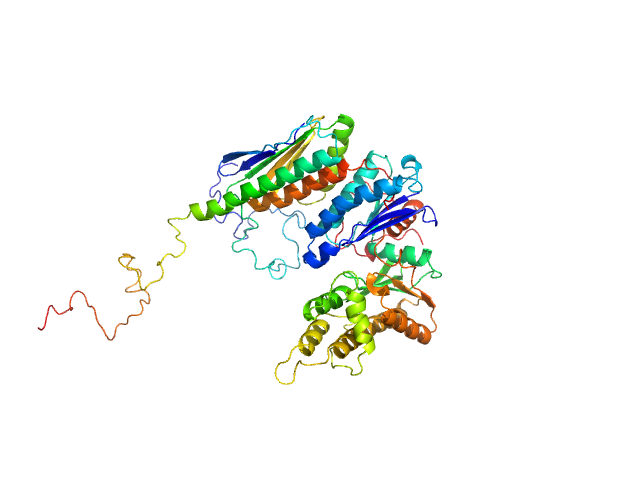
| Sample: |
EKC/KEOPS complex subunit GON7 monomer, 12 kDa Homo sapiens protein
EKC/KEOPS complex subunit LAGE3 monomer, 15 kDa Homo sapiens protein
Probable tRNA N6-adenosine threonylcarbamoyltransferase monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 200 mM NaCl, 5 mM β-mercaptoethanol, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 May 12
|
Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome.
Nat Commun 10(1):3967 (2019)
Arrondel C, Missoury S, Snoek R, Patat J, Menara G, Collinet B, Liger D, Durand D, Gribouval O, Boyer O, Buscara L, Martin G, Machuca E, Nevo F, Lescop E, Braun DA, Boschat AC, Sanquer S, Guerrera IC, Revy P, Parisot M, Masson C, Boddaert N, Charbit M, Decramer S, Novo R, Macher MA, Ranchin B, Bacchetta J, Laurent A, Collardeau-Frachon S, van Eerde AM, Hildebrandt F, Magen D, Antignac C, van Tilbeurgh H, Mollet G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
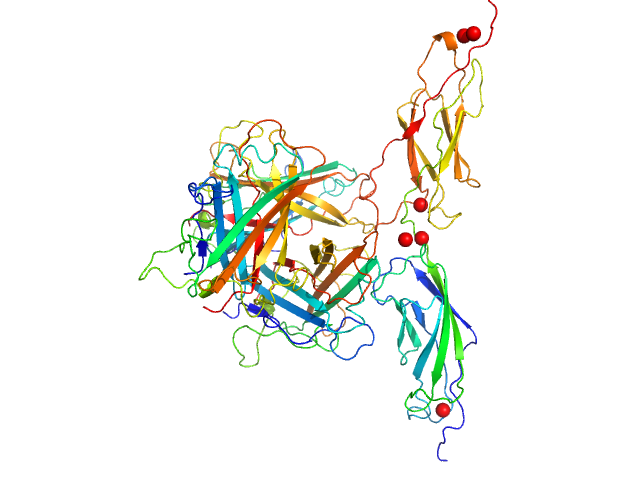
| Sample: |
Human adenovirus serotype 3 fibre knob trimer, 69 kDa Human adenovirus serotype … protein
Human desmoglein 2 extracellular domain 2 and 3 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 3 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jan 26
|
Intermediate-resolution crystal structure of the human adenovirus B serotype 3 fibre knob in complex with the EC2-EC3 fragment of desmoglein 2.
Acta Crystallogr F Struct Biol Commun 75(Pt 12):750-757 (2019)
Vassal-Stermann E, Hutin S, Fender P, Burmeister WP
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|
|
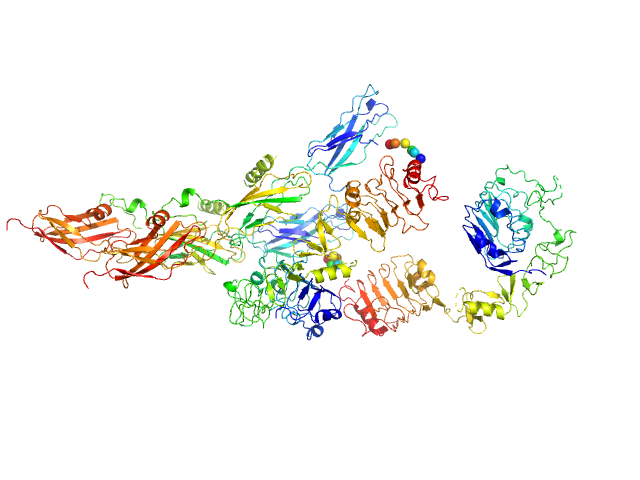
| Sample: |
Insulin receptor-related protein dimer, 201 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 22
|
The dimeric ectodomain of the alkali-sensing insulin receptor-related receptor (ectoIRR) has a droplike shape.
J Biol Chem 294(47):17790-17798 (2019)
Shtykova EV, Petoukhov MV, Mozhaev AA, Deyev IE, Dadinova LA, Loshkarev NA, Goryashchenko AS, Bocharov EV, Jeffries CM, Svergun DI, Batishchev OV, Petrenko AG
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
444 |
nm3 |
|
|
|
|
|
|
|
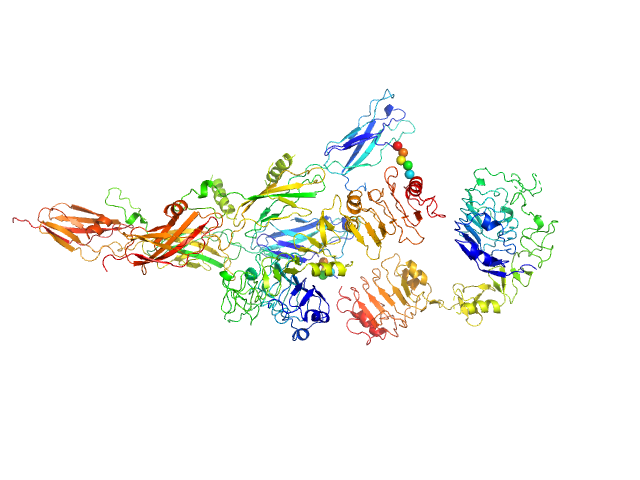
| Sample: |
Insulin receptor-related protein dimer, 201 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 22
|
The dimeric ectodomain of the alkali-sensing insulin receptor-related receptor (ectoIRR) has a droplike shape.
J Biol Chem 294(47):17790-17798 (2019)
Shtykova EV, Petoukhov MV, Mozhaev AA, Deyev IE, Dadinova LA, Loshkarev NA, Goryashchenko AS, Bocharov EV, Jeffries CM, Svergun DI, Batishchev OV, Petrenko AG
|
| RgGuinier |
5.3 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
430 |
nm3 |
|
|
|
|
|
|
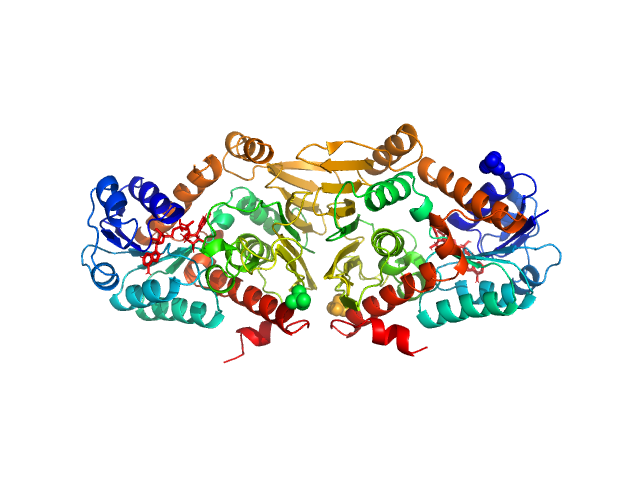
|
| Sample: |
Escherichia coli YjhC dimer, 86 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, 5 % (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 12
|
On the structure and function of Escherichia coli YjhC: an oxidoreductase involved in bacterial sialic acid metabolism.
Proteins (2019)
Horne CR, Kind L, Davies JS, Dobson RCJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
130 |
nm3 |
|
|