|
|
|
|
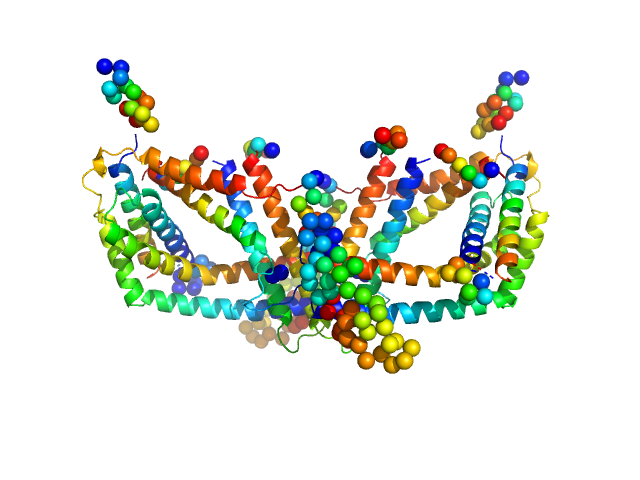
|
| Sample: |
Polyphosphate-targeting protein A dimer, 79 kDa Streptomyces chartreusis protein
|
| Buffer: |
20 mM Tris-HCl 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 24
|
Structural and biochemical analysis of a phosin from Streptomyces chartreusis reveals a combined polyphosphate- and metal-binding fold.
FEBS Lett (2019)
Werten S, Rustmeier NH, Gemmer M, Virolle MJ, Hinrichs W
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
|
|
|
|
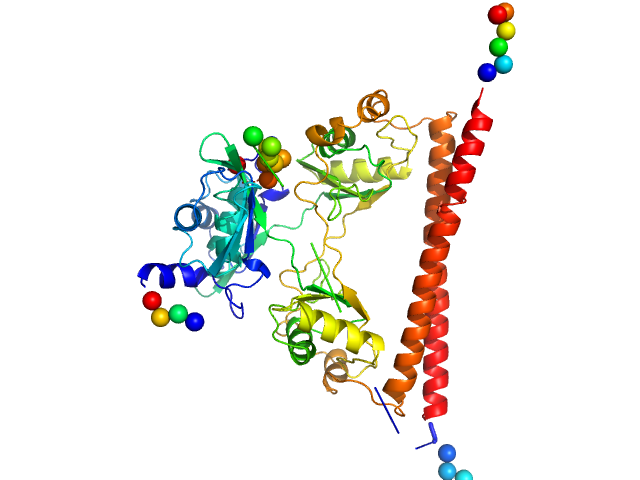
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 250 mM NaCl, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 19
|
A new crystal structure and small-angle X-ray scattering analysis of the homodimer of human SFPQ.
Acta Crystallogr F Struct Biol Commun 75(Pt 6):439-449 (2019)
Hewage TW, Caria S, Lee M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
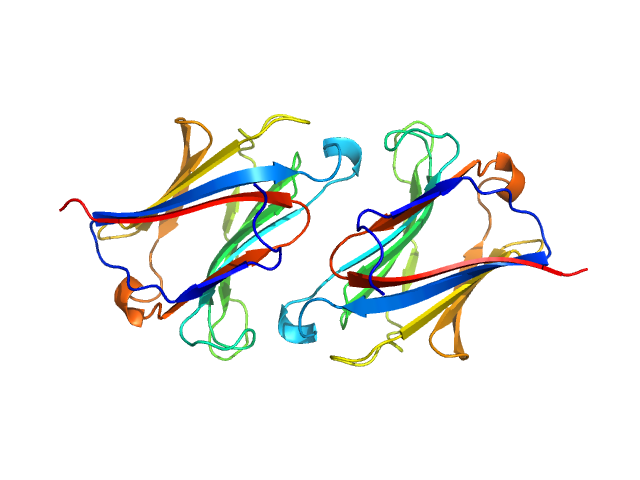
|
| Sample: |
Galectin-10 Tyr69Glu dimer, 33 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes 150 NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Feb 4
|
Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science 364(6442) (2019)
Persson EK, Verstraete K, Heyndrickx I, Gevaert E, Aegerter H, Percier JM, Deswarte K, Verschueren KHG, Dansercoer A, Gras D, Chanez P, Bachert C, Gonçalves A, Van Gorp H, De Haard H, Blanchetot C, Saunders M, Hammad H, Savvides SN, Lambrecht BN
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
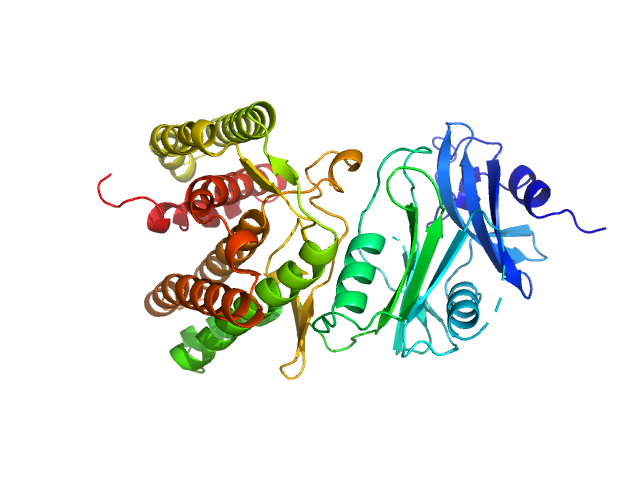
|
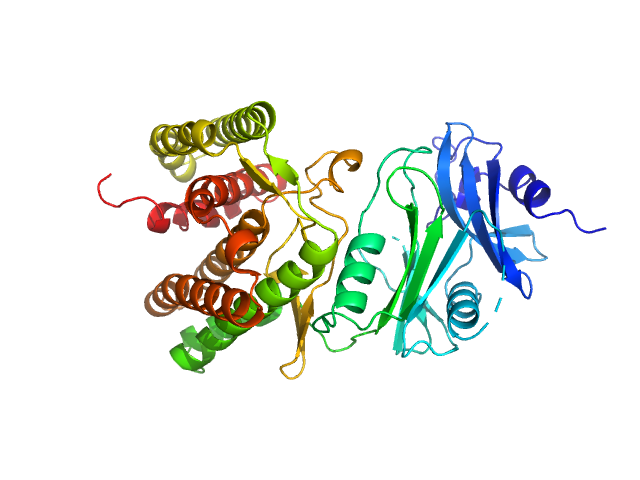
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
|
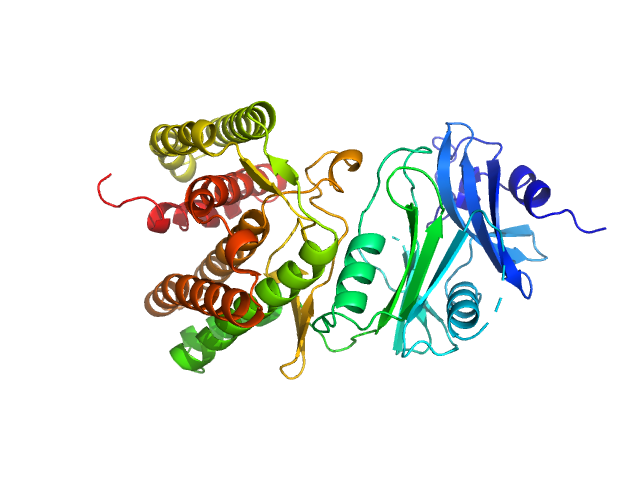
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 0.2 M D-glucosamine, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 20
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
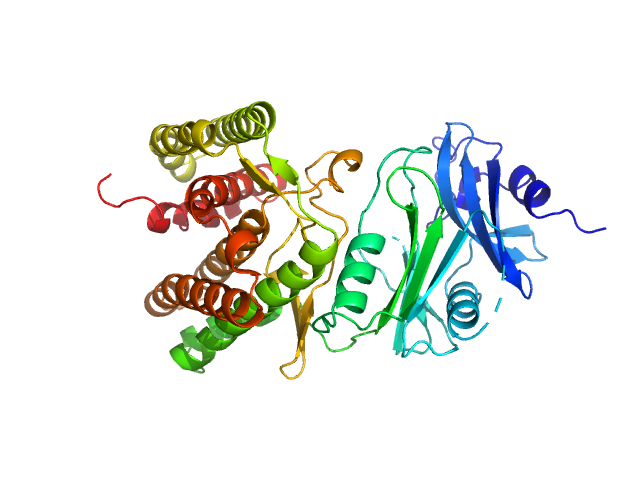
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
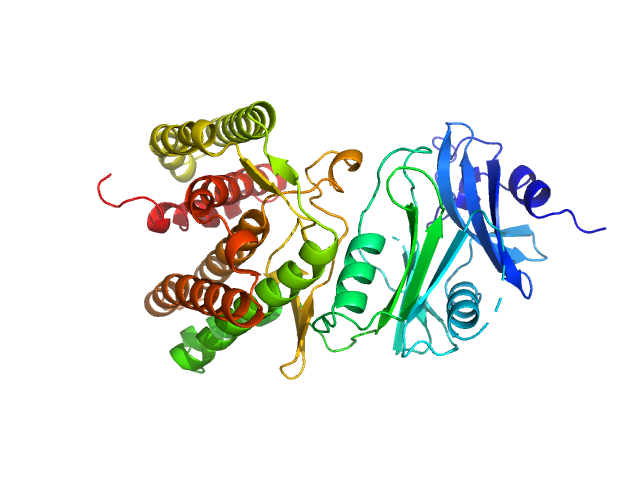
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 0.2 M D-glucosamine, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 20
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
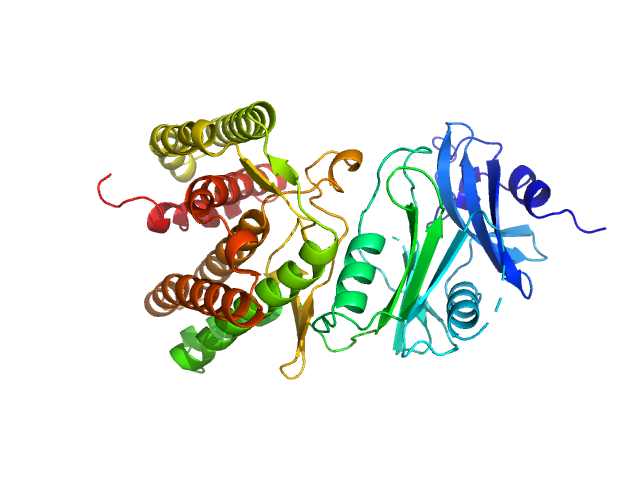
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM D-glucose, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Apr 13
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM D-glucose, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
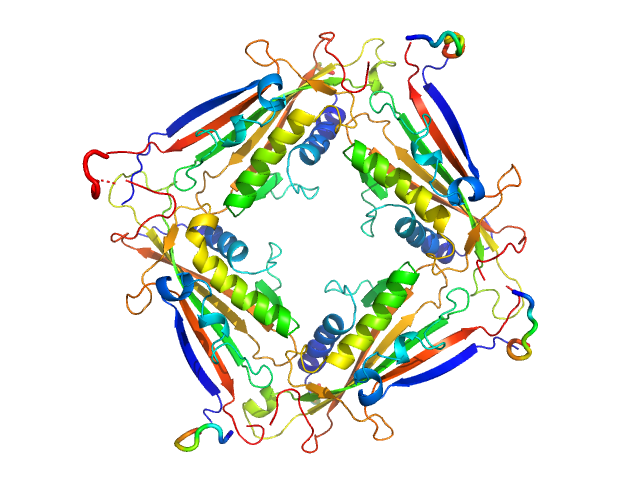
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
134 |
nm3 |
|
|