|
|
|
|
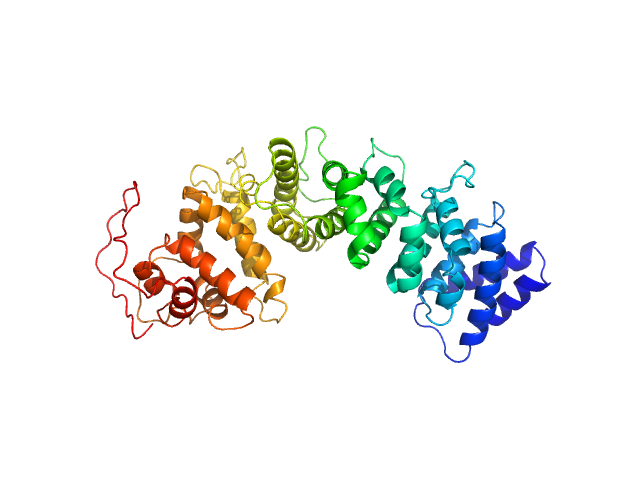
|
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 51 kDa Bos taurus protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 5 % glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Oct 27
|
Structural underpinnings of Ric8A function as a G-protein α-subunit chaperone and guanine-nucleotide exchange factor.
Nat Commun 10(1):3084 (2019)
Srivastava D, Gakhar L, Artemyev NO
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
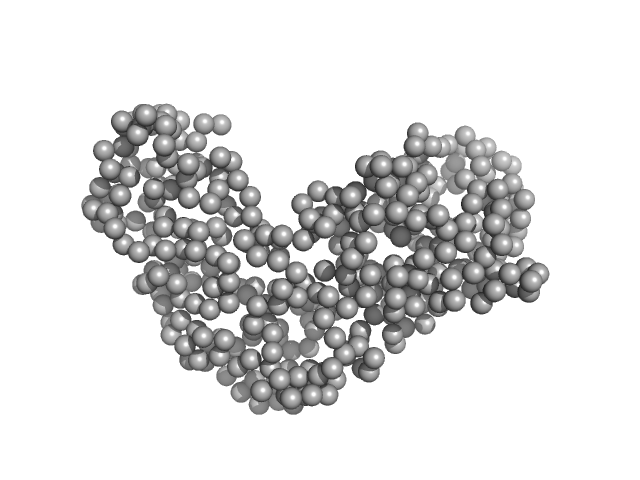
|
| Sample: |
Relaxase (Tra_2) domain of TraI monomer, 46 kDa Neisseria gonorrhoeae protein
|
| Buffer: |
50 mM TRIS-HCl 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 11
|
DNA processing by the MOBH family relaxase TraI encoded within the gonococcal genetic island.
Nucleic Acids Res 47(15):8136-8153 (2019)
Heilers JH, Reiners J, Heller EM, Golzer A, Smits SHJ, van der Does C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
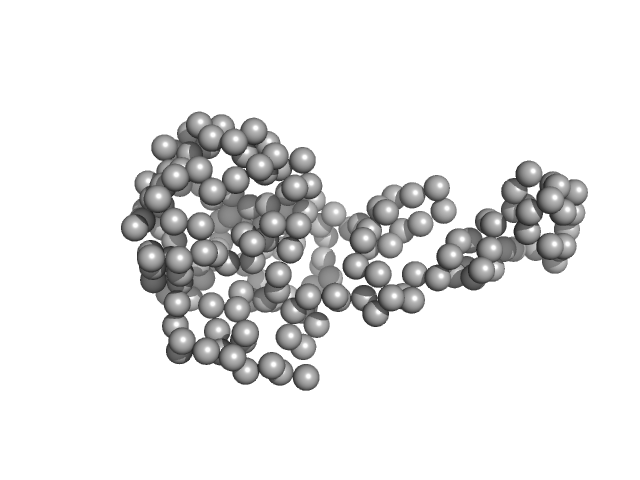
|
| Sample: |
TraI_2_C domain of TraI monomer, 21 kDa Neisseria gonorrhoeae protein
|
| Buffer: |
50 mM TRIS-HCl 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 11
|
DNA processing by the MOBH family relaxase TraI encoded within the gonococcal genetic island.
Nucleic Acids Res 47(15):8136-8153 (2019)
Heilers JH, Reiners J, Heller EM, Golzer A, Smits SHJ, van der Does C
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
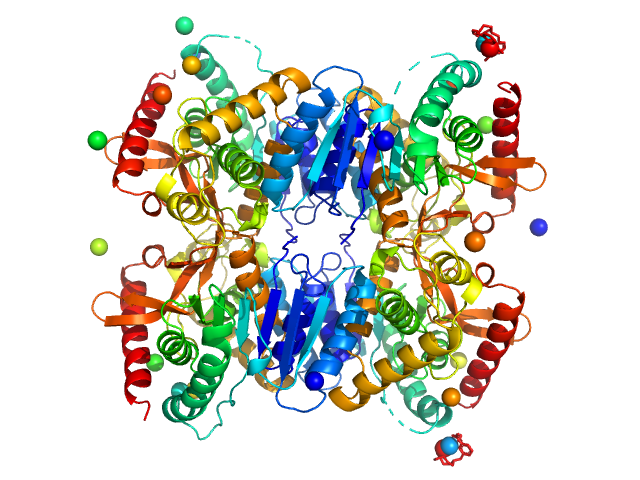
|
| Sample: |
Malate dehydrogenase tetramer, 134 kDa Ignicoccus islandicus DSM … protein
|
| Buffer: |
50 mM Tris-HCl 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 5
|
The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization
Journal of Structural Biology (2019)
Roche J, Girard E, Mas C, Madern D
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|
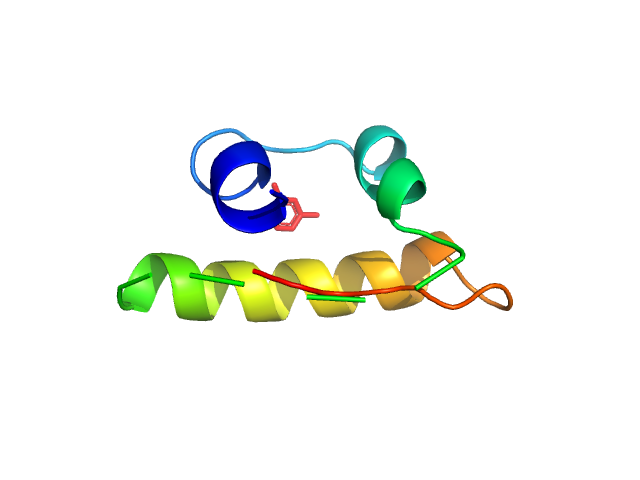
|
| Sample: |
Insulin glulisine hexamer, 35 kDa protein
|
| Buffer: |
Apidra formulation (per ml: 5 mg Sodium chloride, 3.15 mg m-Cresol, 6 mg Trometamol, 0.01 mg Polysorbate 20), pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 20
|
The quaternary structure of insulin glargine and glulisine under formulation conditions.
Biophys Chem 253:106226 (2019)
Nagel N, Graewert MA, Gao M, Heyse W, Jeffries CM, Svergun D, Berchtold H
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.6 |
nm |
|
|
|
|
|
|
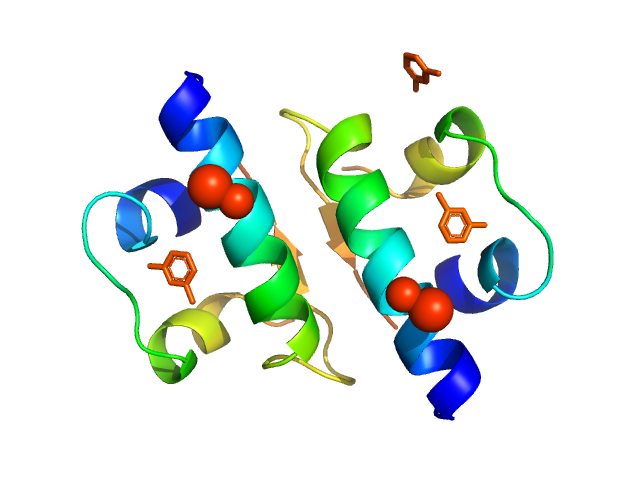
|
| Sample: |
Insulin glargine (Toujeo®) hexamer, 36 kDa protein
|
| Buffer: |
Toujeo Fromulation (190 ug Zinc chloride, 2.7 mg m-Cresol, 20 mg glycerol 85%), pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 5
|
The quaternary structure of insulin glargine and glulisine under formulation conditions.
Biophys Chem 253:106226 (2019)
Nagel N, Graewert MA, Gao M, Heyse W, Jeffries CM, Svergun D, Berchtold H
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
Fatty acid oxidation complex subunit alpha monomer, 81 kDa Escherichia coli protein
Fatty acid oxidation complex subunit alpha monomer, 81 kDa Escherichia coli protein
3-ketoacyl-CoA thiolase FadA (beta subunit) dimer, 82 kDa Escherichia coli protein
|
| Buffer: |
20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 120 mM KCl, 2.5 mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 May 30
|
Complementary substrate specificity and distinct quaternary assembly of the Escherichia coli aerobic and anaerobic beta-oxidation trifunctional enzyme complexes.
Biochem J (2019)
Sah-Teli SK, Hynönen MJ, Schmitz W, Geraets JA, Seitsonen J, Pedersen JS, Butcher SJ, Wierenga RK, Venkatesan R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
406 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli (strain … protein
Anaerobic Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli protein
Anaerobic Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli protein
Anaerobic Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli protein
Anaerobic 3-ketoacyl-CoA thiolase FadI beta subunit dimer, 96 kDa Escherichia coli protein
Anaerobic 3-ketoacyl-CoA thiolase FadI beta subunit dimer, 96 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 5% glycerol, 0.05% C12E9 (1-O-(n-Dodecyl)-nonaethyleneglycol), 2.5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 22
|
Complementary substrate specificity and distinct quaternary assembly of the Escherichia coli aerobic and anaerobic beta-oxidation trifunctional enzyme complexes.
Biochem J (2019)
Sah-Teli SK, Hynönen MJ, Schmitz W, Geraets JA, Seitsonen J, Pedersen JS, Butcher SJ, Wierenga RK, Venkatesan R
|
| RgGuinier |
6.2 |
nm |
| Dmax |
19.6 |
nm |
| VolumePorod |
856 |
nm3 |
|
|
|
|
|
|
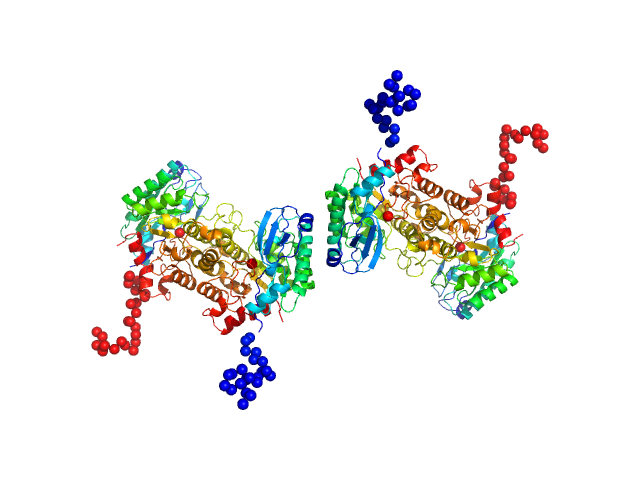
|
| Sample: |
2-amino-3-carboxymuconate 6-semialdehyde decarboxylase tetramer, 159 kDa Pseudomonas fluorescens protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Jul 15
|
Quaternary structure of α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase (ACMSD) controls its activity.
J Biol Chem 294(30):11609-11621 (2019)
Yang Y, Davis I, Matsui T, Rubalcava I, Liu A
|
| RgGuinier |
5.2 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
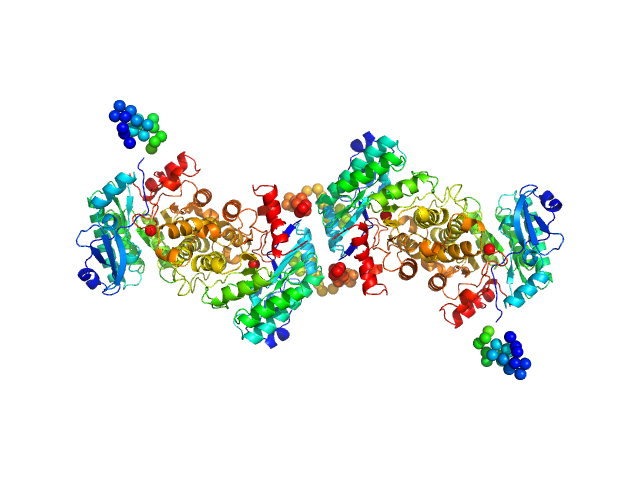
|
| Sample: |
2-amino-3-carboxymuconate 6-semialdehyde decarboxylase tetramer, 159 kDa Pseudomonas fluorescens protein
|
| Buffer: |
25 mM HEPES, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Jan 10
|
Quaternary structure of α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase (ACMSD) controls its activity.
J Biol Chem 294(30):11609-11621 (2019)
Yang Y, Davis I, Matsui T, Rubalcava I, Liu A
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
195 |
nm3 |
|
|