|
|
|
|
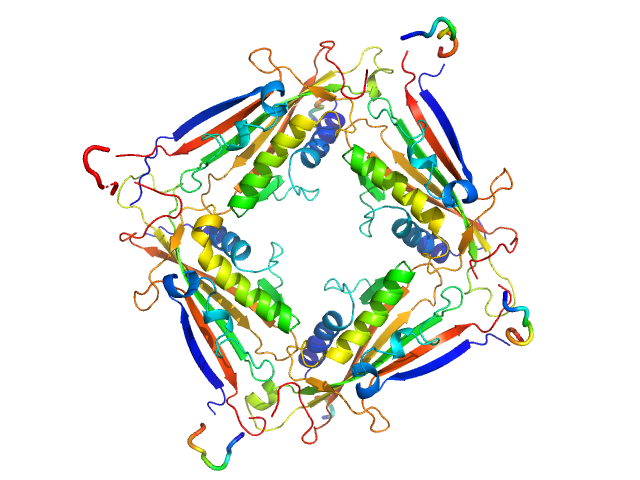
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 7.4, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
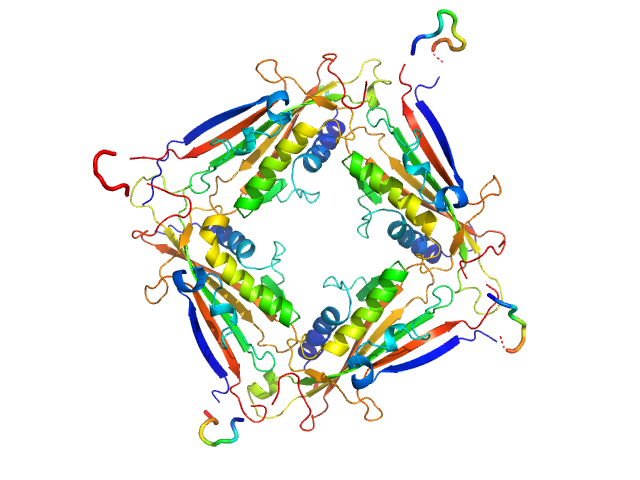
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 6.5, 150 mM NaCl, 0.5 mM CaCl2, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
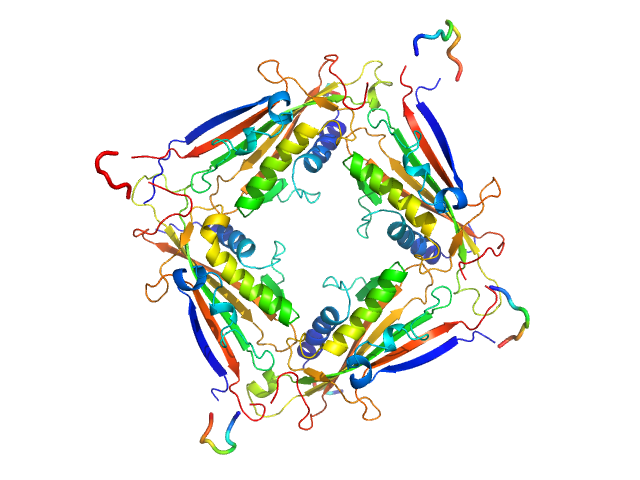
|
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 6.5 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
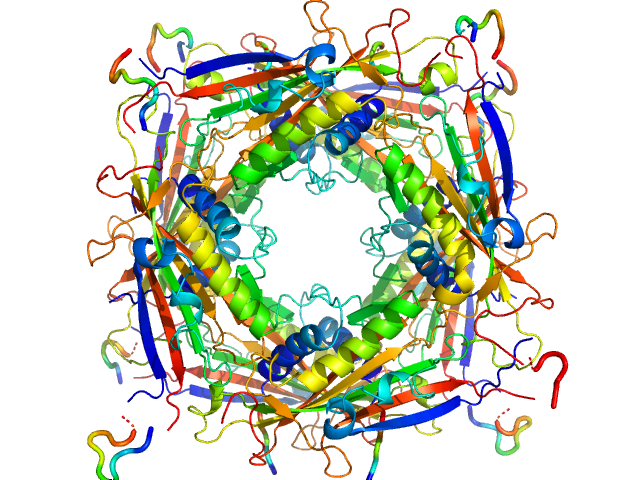
|
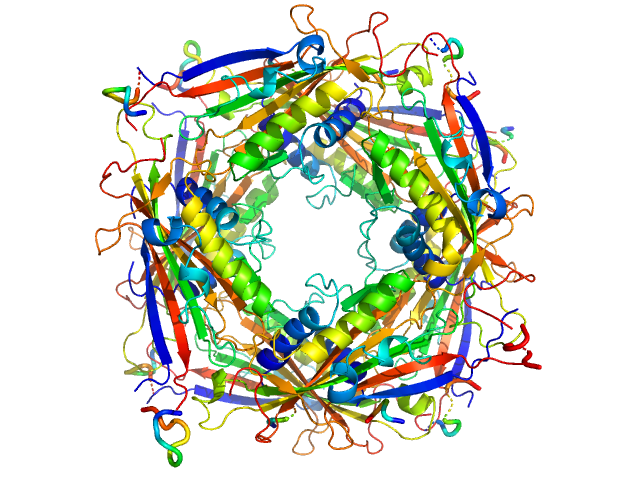
| Sample: |
Transient receptor potential channel mucolipin 2 octamer, 187 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 4.5, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
285 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential channel mucolipin 2 octamer, 187 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 4.5, 150 mM NaCl, 0.5 mM CaCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
|
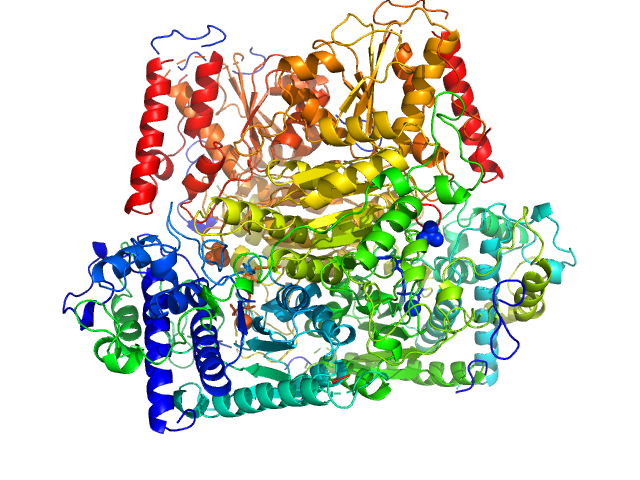
| Sample: |
Probable phosphoketolase dimer, 191 kDa Lactococcus lactis subsp. … protein
|
| Buffer: |
20mM potassium phosphate 150mM NaCl 0.007 %(w/v) β-octyl glucoside 1mM DTT 1mM MgCl 1mM thiaminpyrophosphate 2 %(v/v) glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 7
|
Crystal structure of a xylulose 5-phosphate phosphoketolase. insights into the substrate specificity for xylulose 5-phosphate.
J Struct Biol (2019)
Scheidig AJ, Horvath D, Szedlacsek SE
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
|
|
|
|
|
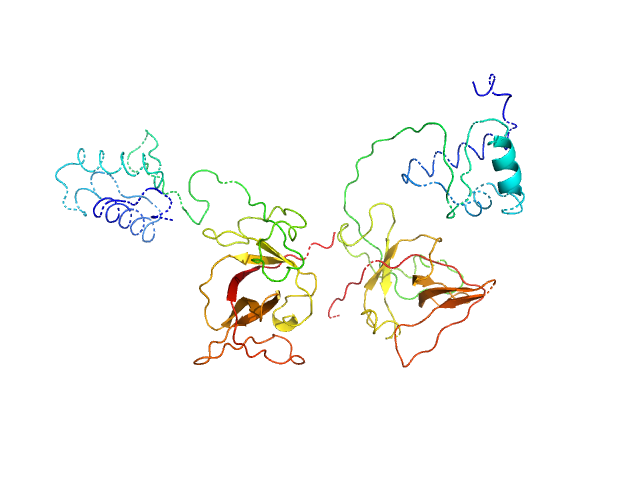
| Sample: |
Bacillus thuringiensis LexA repressor dimer, 47 kDa Bacillus thuringiensis protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 10% glycerol,, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Aug 25
|
Structural Insights into Bacteriophage GIL01 gp7 Inhibition of Host LexA Repressor.
Structure 27(7):1094-1102.e4 (2019)
Caveney NA, Pavlin A, Caballero G, Bahun M, Hodnik V, de Castro L, Fornelos N, Butala M, Strynadka NCJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
|
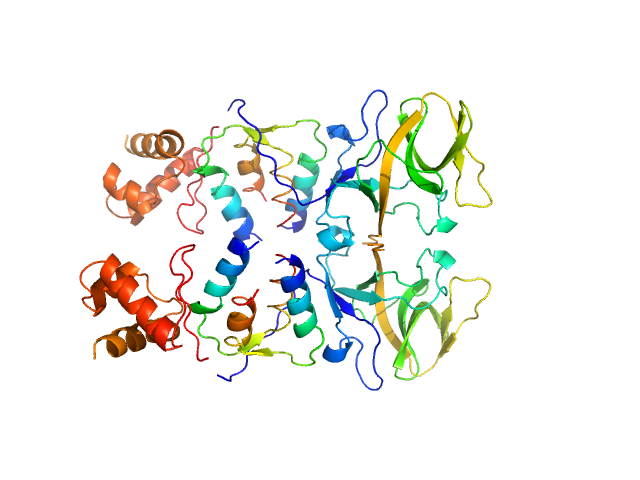
| Sample: |
Bacillus thuringiensis LexA repressor dimer, 47 kDa Bacillus thuringiensis protein
Bacteriophage pGIL01 gp7 tetramer, 24 kDa Bacteriophage pGIL01 protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 10% glycerol,, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Aug 25
|
Structural Insights into Bacteriophage GIL01 gp7 Inhibition of Host LexA Repressor.
Structure 27(7):1094-1102.e4 (2019)
Caveney NA, Pavlin A, Caballero G, Bahun M, Hodnik V, de Castro L, Fornelos N, Butala M, Strynadka NCJ
|
|
|
|
|
|
|
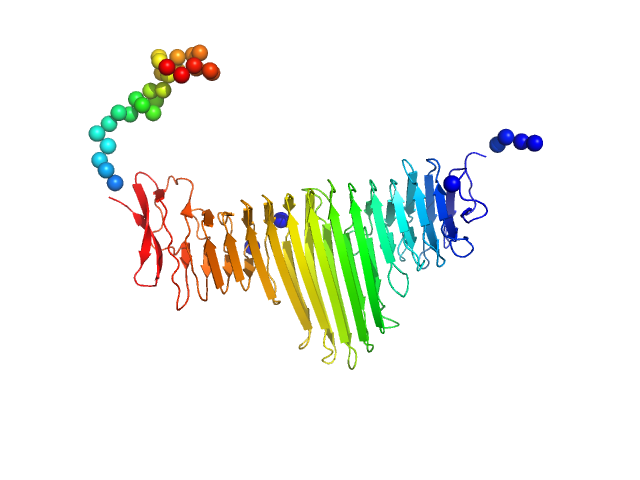
|
| Sample: |
Alpha domain of autotransporter protein UpaB monomer, 48 kDa E. Coli CFT073 protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 May 1
|
Unique structural features of a bacterial autotransporter adhesin suggest mechanisms for interaction with host macromolecules.
Nat Commun 10(1):1967 (2019)
Paxman JJ, Lo AW, Sullivan MJ, Panjikar S, Kuiper M, Whitten AE, Wang G, Luan CH, Moriel DG, Tan L, Peters KM, Phan MD, Gee CL, Ulett GC, Schembri MA, Heras B
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
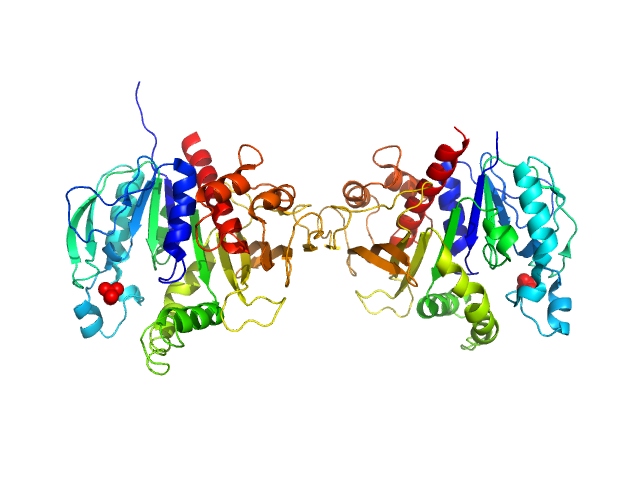
|
| Sample: |
Phosphoribulokinase, chloroplastic dimer, 78 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
Tris-HCl 50 mM 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Arabidopsis and Chlamydomonas phosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc Natl Acad Sci U S A 116(16):8048-8053 (2019)
Gurrieri L, Del Giudice A, Demitri N, Falini G, Pavel NV, Zaffagnini M, Polentarutti M, Crozet P, Marchand CH, Henri J, Trost P, Lemaire SD, Sparla F, Fermani S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
115 |
nm3 |
|
|