|
|
|
|
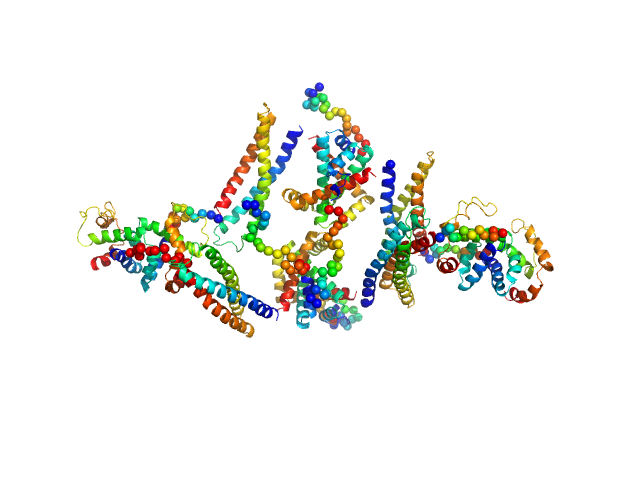
|
| Sample: |
HP2042 form from Helicobacter pylori, N-terminal domain of syntaxin-1A from Rattus norvegicus, Trp repressor from Escherichia coli tetramer, 150 kDa Helicobacter pylori, Rattus … protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku Nano-Viewer, Nara Institute of Science and Technology on 2016 Dec 8
|
Construction of a Quadrangular Tetramer and a Cage-Like Hexamer from Three-Helix Bundle-Linked Fusion Proteins.
ACS Synth Biol (2019)
Miyamoto T, Hayashi Y, Yoshida K, Watanabe H, Uchihashi T, Yonezawa K, Shimizu N, Kamikubo H, Hirota S
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
379 |
nm3 |
|
|
|
|
|
|
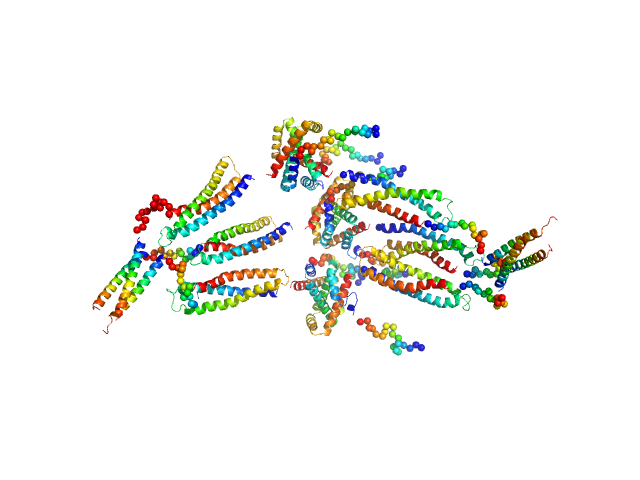
|
| Sample: |
HP0242 from Helicobacter pylori, N-terminal domain of syntaxin-1A from Rattus norvegicus, de novo designed coiled-coil trimer domain hexamer, 179 kDa Helicobacter pylori, Rattus … protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku Nano-Viewer, Nara Institute of Science and Technology on 2017 Mar 29
|
Construction of a Quadrangular Tetramer and a Cage-Like Hexamer from Three-Helix Bundle-Linked Fusion Proteins.
ACS Synth Biol (2019)
Miyamoto T, Hayashi Y, Yoshida K, Watanabe H, Uchihashi T, Yonezawa K, Shimizu N, Kamikubo H, Hirota S
|
| RgGuinier |
6.5 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
642 |
nm3 |
|
|
|
|
|
|
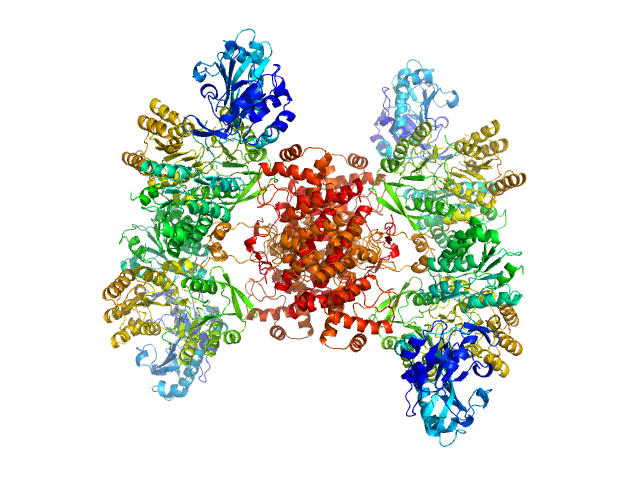
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 6
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.0 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
738 |
nm3 |
|
|
|
|
|
|
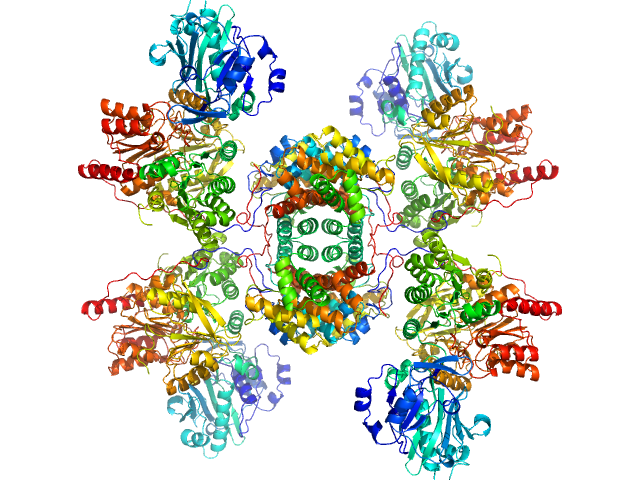
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 5
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
5.8 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
709 |
nm3 |
|
|
|
|
|
|
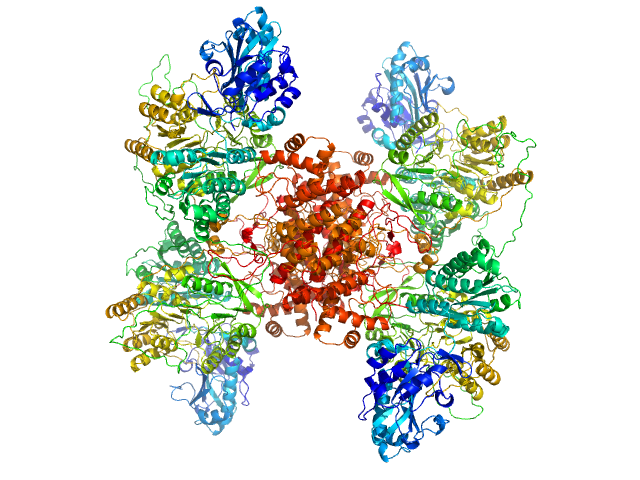
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 4
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
5.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
775 |
nm3 |
|
|
|
|
|
|
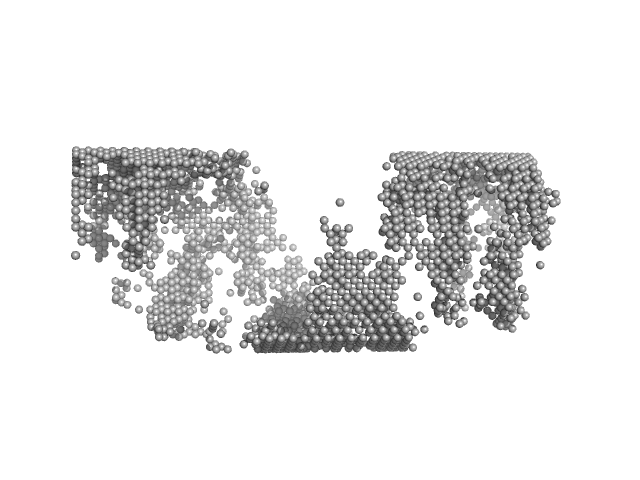
|
| Sample: |
Matrix protein, 40 kDa Newcastle disease virus … protein
|
| Buffer: |
STE buffer 100 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 11
|
Solution Structure, Self-Assembly, and Membrane Interactions of the Matrix Protein from Newcastle Disease Virus at Neutral and Acidic pH
Journal of Virology 93(6) (2019)
Shtykova E, Petoukhov M, Dadinova L, Fedorova N, Tashkin V, Timofeeva T, Ksenofontov A, Loshkarev N, Baratova L, Jeffries C, Svergun D, Batishchev O, García-Sastre A
|
|
|
|
|
|
|
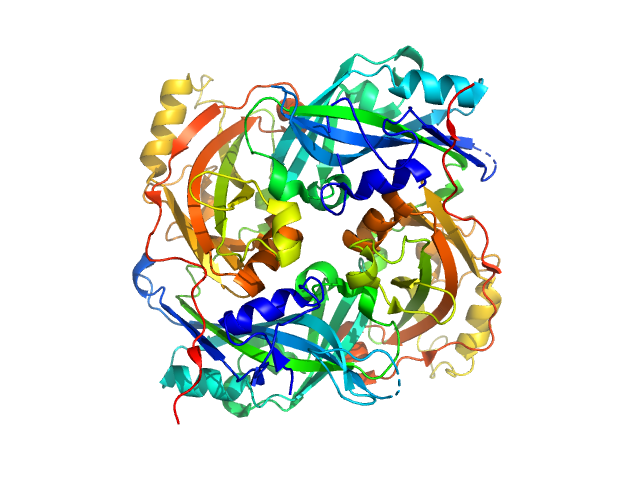
|
| Sample: |
Matrix protein dimer, 79 kDa Newcastle disease virus … protein
|
| Buffer: |
STE buffer 100 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH: 4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 11
|
Solution Structure, Self-Assembly, and Membrane Interactions of the Matrix Protein from Newcastle Disease Virus at Neutral and Acidic pH
Journal of Virology 93(6) (2019)
Shtykova E, Petoukhov M, Dadinova L, Fedorova N, Tashkin V, Timofeeva T, Ksenofontov A, Loshkarev N, Baratova L, Jeffries C, Svergun D, Batishchev O, García-Sastre A
|
|
|
|
|
|
|

|
| Sample: |
DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein) trimer, 81 kDa Wolbachia endosymbiont of … protein
|
| Buffer: |
25 mM TRIS, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2014 Mar 29
|
The atypical thiol-disulfide exchange protein α-DsbA2 from Wolbachia pipientis is a homotrimeric disulfide isomerase.
Acta Crystallogr D Struct Biol 75(Pt 3):283-295 (2019)
Walden PM, Whitten AE, Premkumar L, Halili MA, Heras B, King GJ, Martin JL
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
913 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DsbA-like disulfide oxidoreductase (thiol-disulfide exchange protein) monomer, 21 kDa Wolbachia endosymbiont of … protein
|
| Buffer: |
25 mM TRIS, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2012 Feb 29
|
The atypical thiol-disulfide exchange protein α-DsbA2 from Wolbachia pipientis is a homotrimeric disulfide isomerase.
Acta Crystallogr D Struct Biol 75(Pt 3):283-295 (2019)
Walden PM, Whitten AE, Premkumar L, Halili MA, Heras B, King GJ, Martin JL
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|
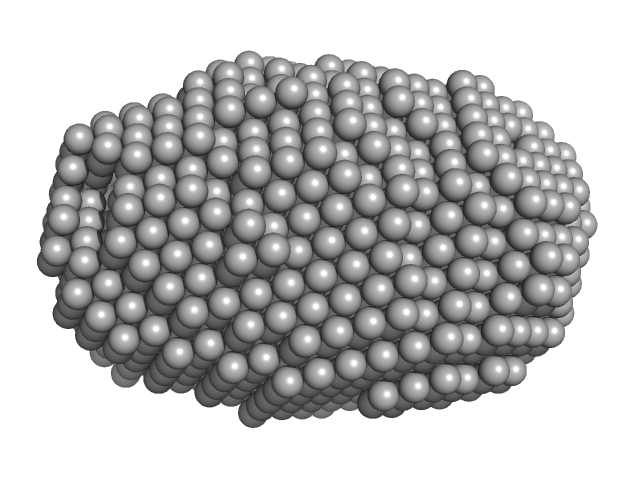
|
| Sample: |
C-terminal catalytic domain of Suppressor of Copper Sensitivity C protein monomer, 20 kDa Proteus mirabilis protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 5
|
Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol 75(Pt 3):296-307 (2019)
Furlong EJ, Kurth F, Premkumar L, Whitten AE, Martin JL
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
22200 |
nm3 |
|
|