|
|
|
|
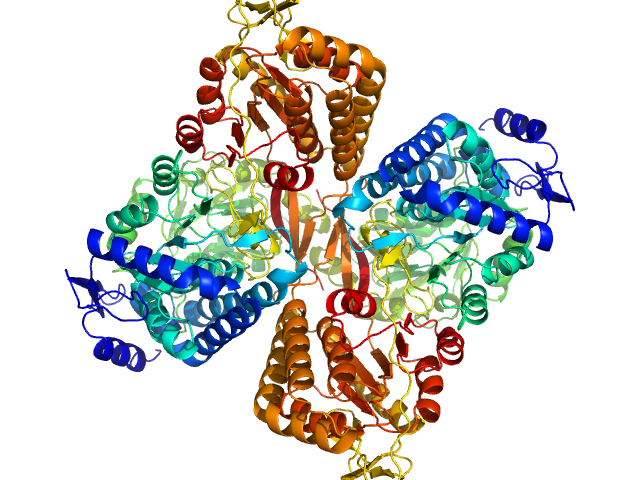
|
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|
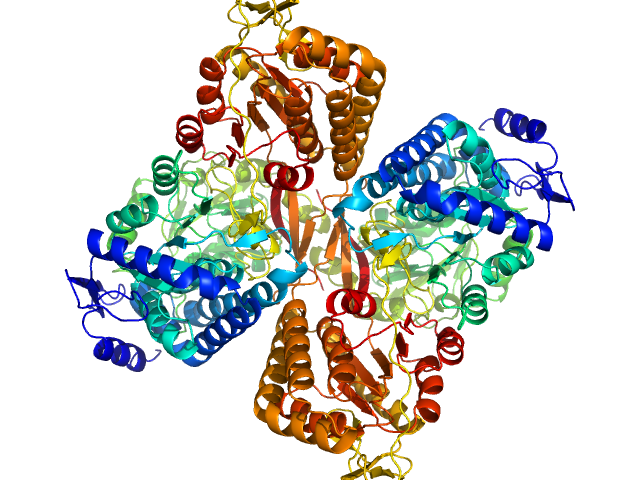
|
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
205 |
nm3 |
|
|
|
|
|
|
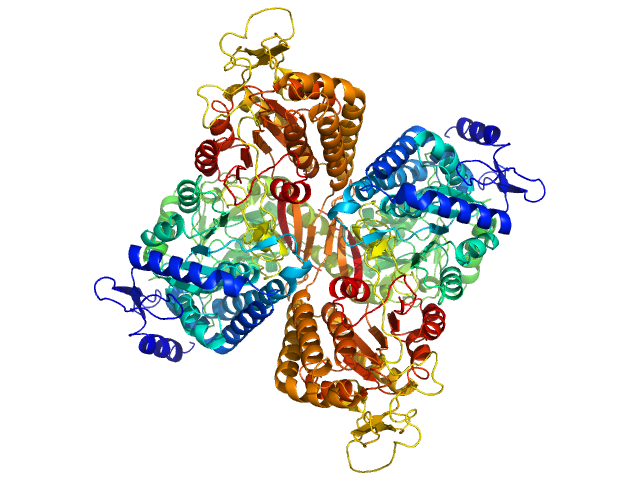
|
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
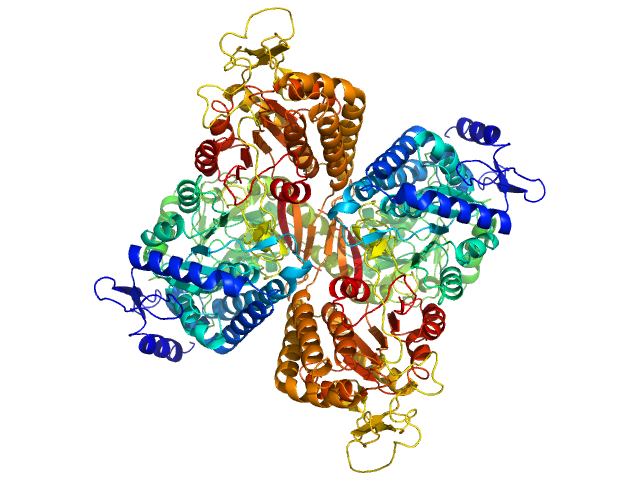
|
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
|
|
|
|
|
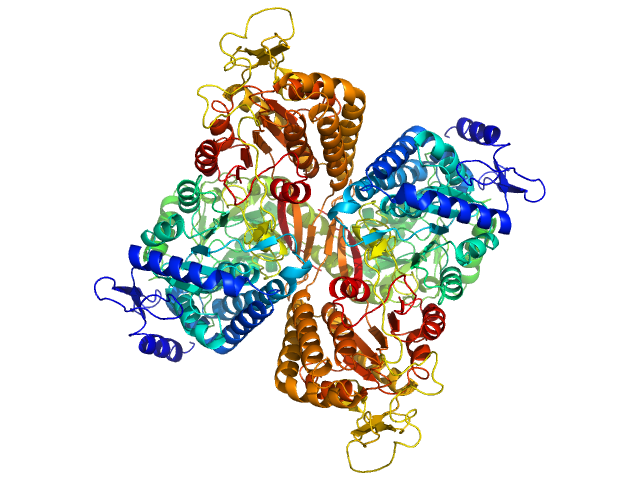
| Sample: |
Aldehyde dehydrogenase family 16 member A1 from Homo sapiens dimer, 171 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
|
|
|
|
|
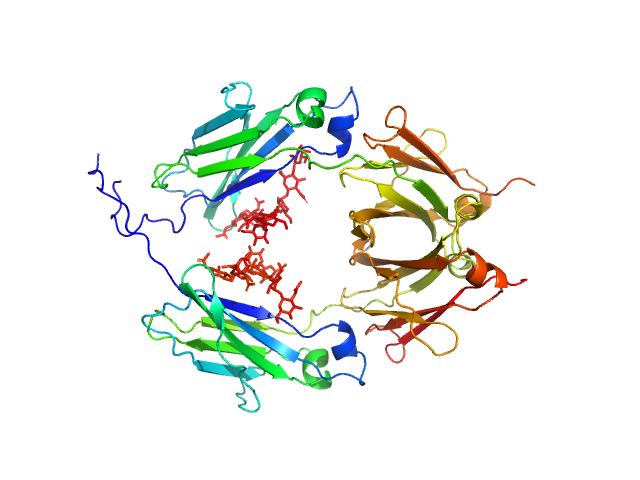
| Sample: |
Glycosylated human immunoglobulin G Fc region dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Citrate-Phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Mar 5
|
CH2 domain orientation of human immunoglobulin G in solution: Structural comparison of glycosylated and aglycosylated Fc regions using small-angle X-ray scattering.
MAbs (2018)
Yageta S, Imamura H, Shibuya R, Honda S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
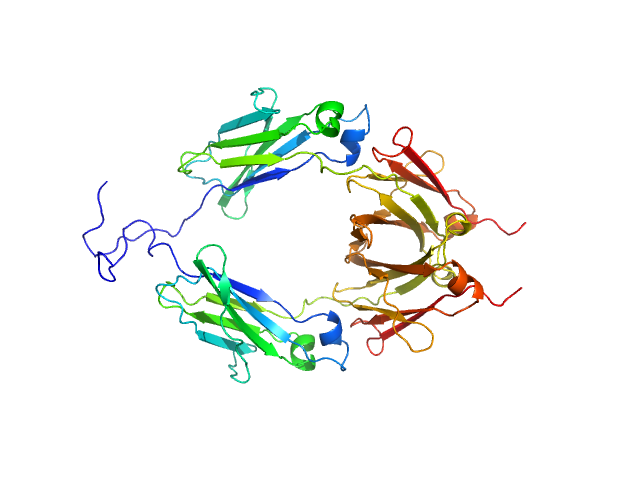
| Sample: |
Aglycosylated human immunoglobulin G Fc region dimer, 51 kDa Homo sapiens protein
|
| Buffer: |
20 mM Citrate-Phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Mar 5
|
CH2 domain orientation of human immunoglobulin G in solution: Structural comparison of glycosylated and aglycosylated Fc regions using small-angle X-ray scattering.
MAbs (2018)
Yageta S, Imamura H, Shibuya R, Honda S
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
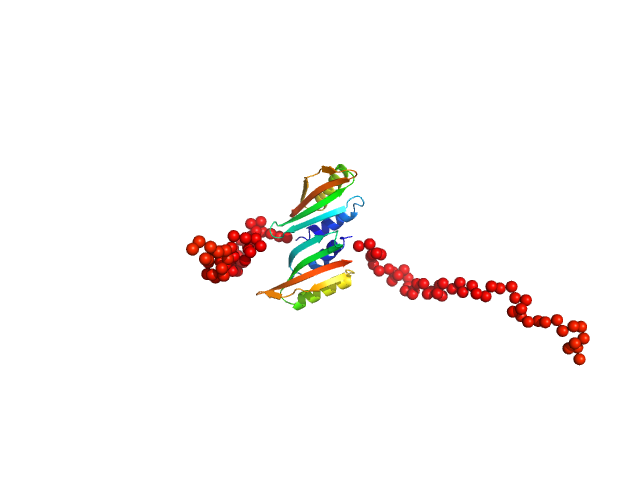
| Sample: |
Type II secretion system protein L, periplasmic domain dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
50 mM TRIS, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 8
|
Structure and oligomerization of the periplasmic domain of GspL from the type II secretion system of Pseudomonas aeruginosa.
Sci Rep 8(1):16760 (2018)
Fulara A, Vandenberghe I, Read RJ, Devreese B, Savvides SN
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
|
|
|
|
|
|
|
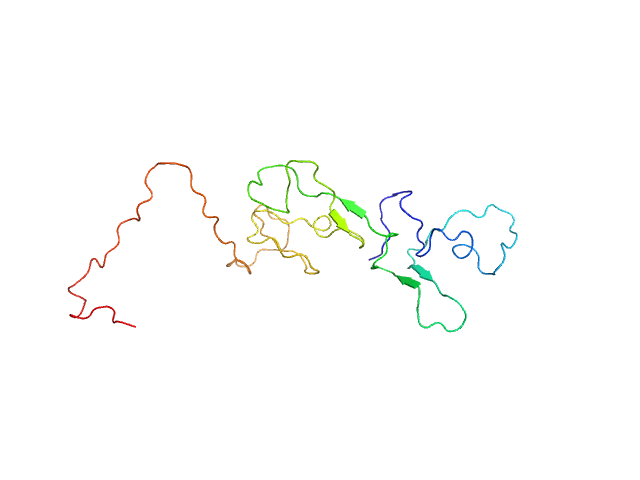
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 0.05 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2017 Jun 12
|
A Metastable Contact and Structural Disorder in the Estrogen Receptor Transactivation Domain.
Structure 27(2):229-240.e4 (2019)
Peng Y, Cao S, Kiselar J, Xiao X, Du Z, Hsieh A, Ko S, Chen Y, Agrawal P, Zheng W, Shi W, Jiang W, Yang L, Chance MR, Surewicz WK, Buck M, Yang S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|
|
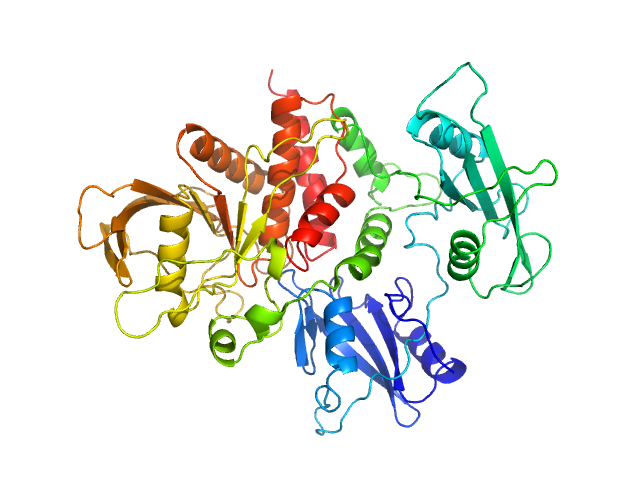
| Sample: |
Tyrosine-protein phosphatase non-receptor type 11 monomer, 61 kDa Homo sapiens protein
|
| Buffer: |
50 mM ADA, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2016 Mar 28
|
Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun 9(1):4507 (2018)
Pádua RAP, Sun Y, Marko I, Pitsawong W, Stiller JB, Otten R, Kern D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
|
|