|
|
|
|
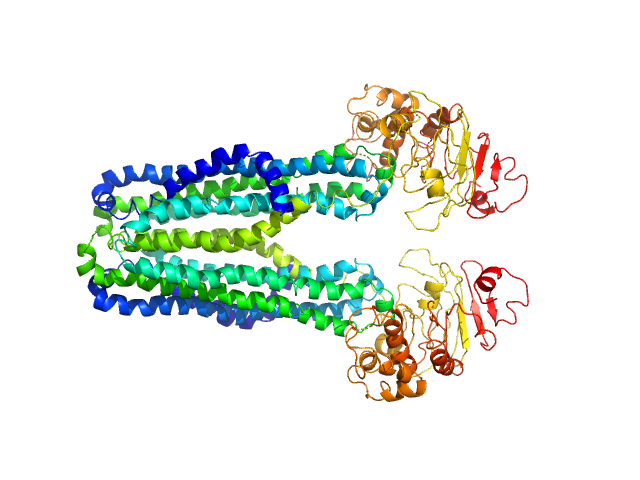
|
| Sample: |
Lipid A export ATP-binding/permease protein MsbA dimer, 133 kDa Escherichia coli protein
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at D11, ILL on 2017 Mar 9
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
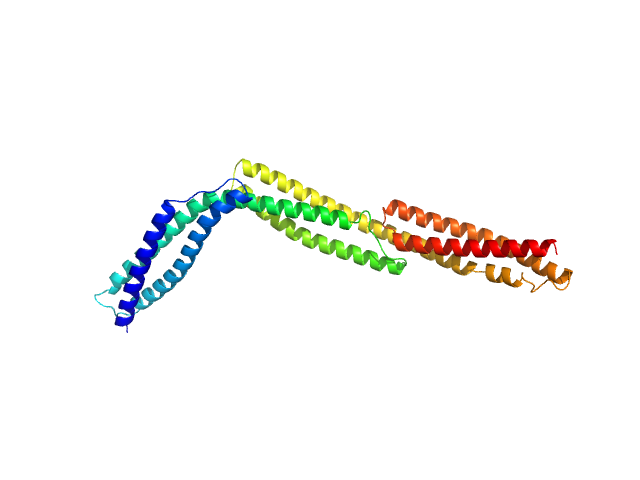
|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
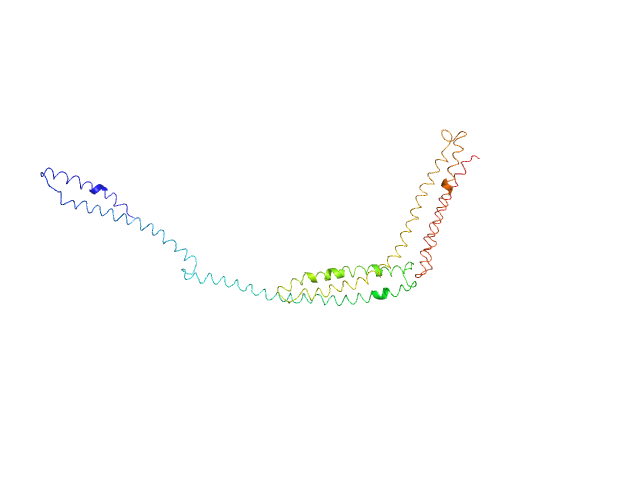
|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
6.2 |
nm |
| Dmax |
24.8 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
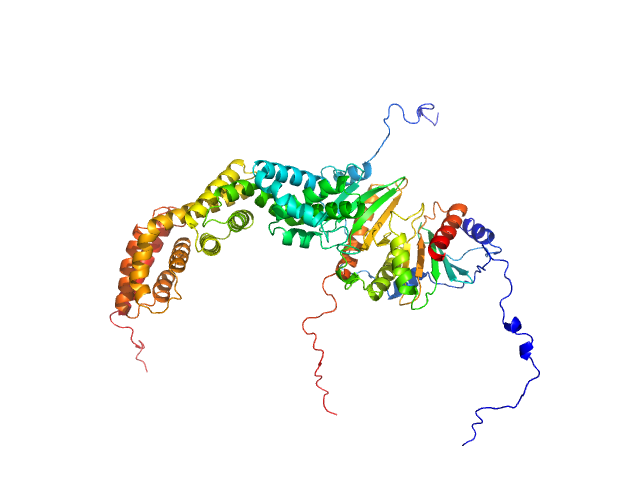
|
| Sample: |
Type III secretion protein HrpG, 19 kDa Erwinia amylovora protein
Type III secretion protein HrpV, 13 kDa Erwinia amylovora protein
Hypersensitivity response secretion protein HrpJ, 42 kDa Erwinia amylovora protein
|
| Buffer: |
20mM Tris-Cl, pH 8.0, 100mM NaCl, 2mM DTT, 0.5mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 3
|
Migration of Type III Secretion System Transcriptional Regulators Links Gene Expression to Secretion.
MBio 9(4) (2018)
Charova SN, Gazi AD, Mylonas E, Pozidis C, Sabarit B, Anagnostou D, Psatha K, Aivaliotis M, Beuzon CR, Panopoulos NJ, Kokkinidis M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
|
|
|
|
|
|

|
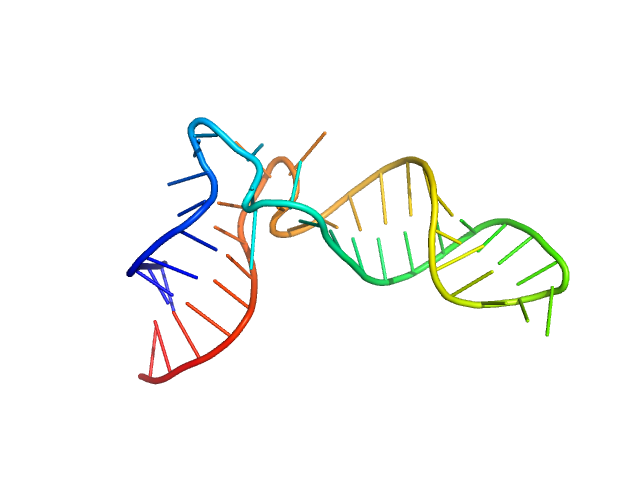
| Sample: |
Arginyl transfer RNA monomer, 13 kDa Romanomermis culicivorax RNA
|
| Buffer: |
50 mM HEPES-NaOH 10 mM MgCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Dec 16
|
Small but large enough: structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax.
Nucleic Acids Res (2018)
Jühling T, Duchardt-Ferner E, Bonin S, Wöhnert J, Pütz J, Florentz C, Betat H, Sauter C, Mörl M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoleucyl transfer RNA monomer, 15 kDa Romanomermis culicivorax RNA
|
| Buffer: |
50 mM HEPES-NaOH 10 mM MgCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Dec 16
|
Small but large enough: structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax.
Nucleic Acids Res (2018)
Jühling T, Duchardt-Ferner E, Bonin S, Wöhnert J, Pütz J, Florentz C, Betat H, Sauter C, Mörl M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Designed Ankyrin Repeat Protein D1 monomer, 18 kDa synthetic construct protein
Kinesin-like protein KIF2A monomer, 48 kDa Homo sapiens protein
Tubulin alpha-1B chain dimer, 100 kDa Bos taurus protein
Tubulin beta-2B chain dimer, 100 kDa Bos taurus protein
|
| Buffer: |
HEPES 20 mM, MgCl2 1mM, NaCl 150mM, pH: 7.2 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 16
|
Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate.
Nat Commun 9(1):2628 (2018)
Trofimova D, Paydar M, Zara A, Talje L, Kwok BH, Allingham JS
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
374 |
nm3 |
|
|
|
|
|
|
|
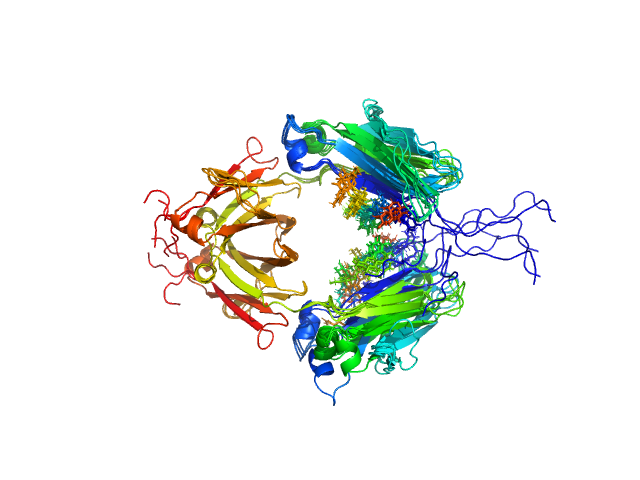
| Sample: |
Immunoglobulin heavy constant gamma 1 dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
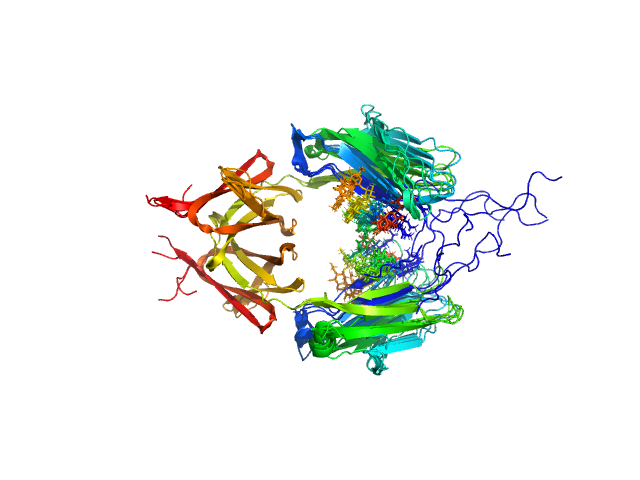
| Sample: |
Immunoglobulin heavy constant gamma 2 dimer, 52 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
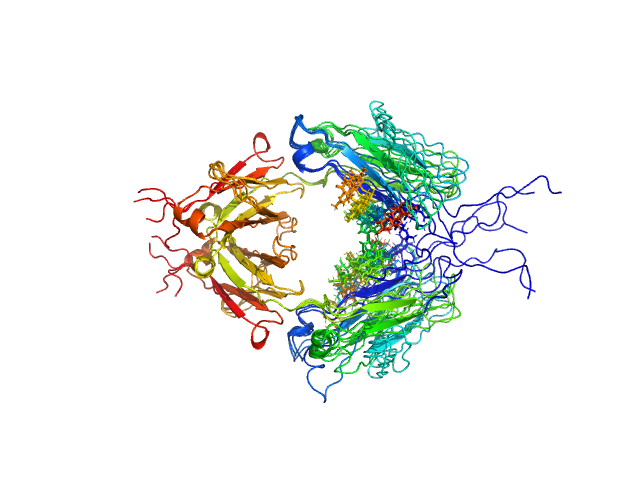
| Sample: |
Immunoglobulin heavy constant gamma 1 M255Y/S257T/T259E dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|