|
|
|
|
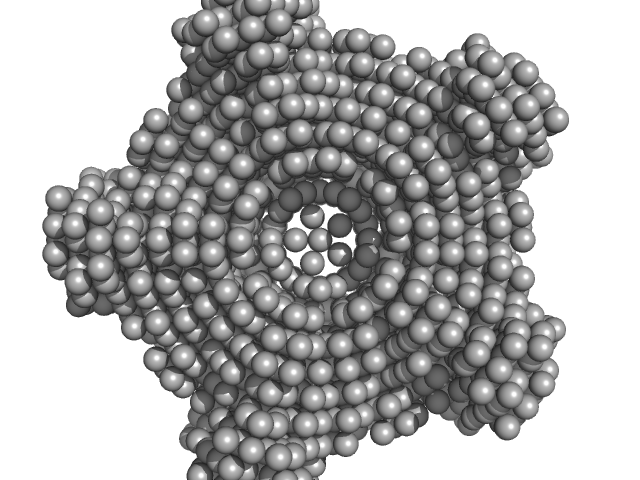
|
| Sample: |
Class I fructose-1,6-bisphosphate aldolase (FbaB) from E. coli decamer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 30
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
484 |
nm3 |
|
|
|
|
|
|
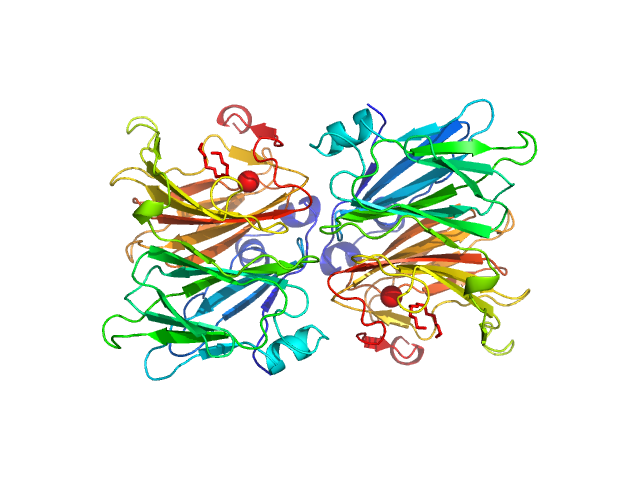
|
| Sample: |
5-keto-4-deoxyuronate isomerase (KduI) from E. coli None, Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 20
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
|
|
|
|
|
|
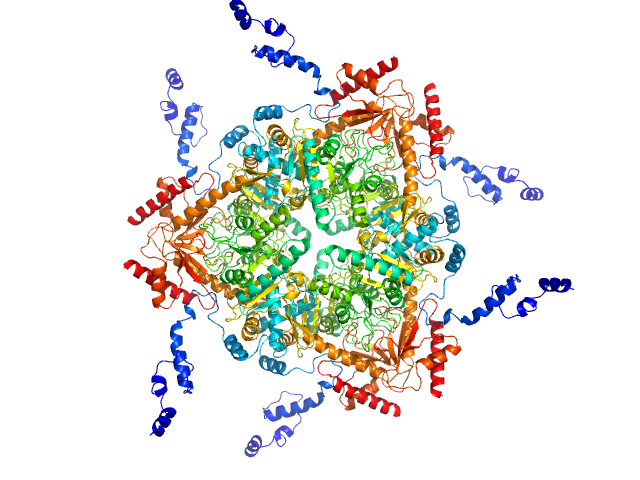
|
| Sample: |
Glutamate decarboxylase alpha (GadA) from E. coli monomer, 53 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 20
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.8 |
nm |
| VolumePorod |
410 |
nm3 |
|
|
|
|
|
|
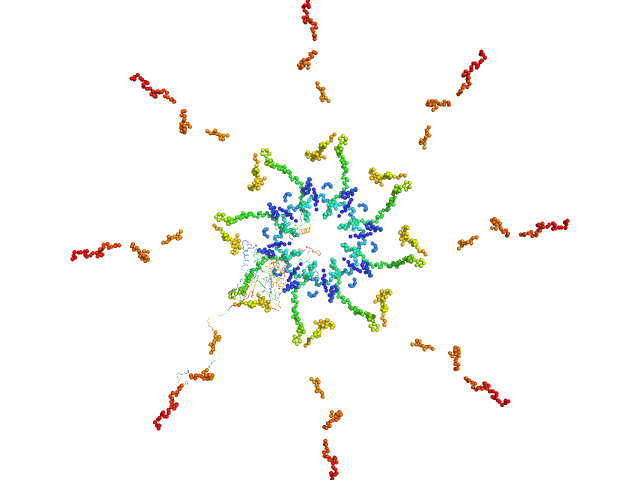
|
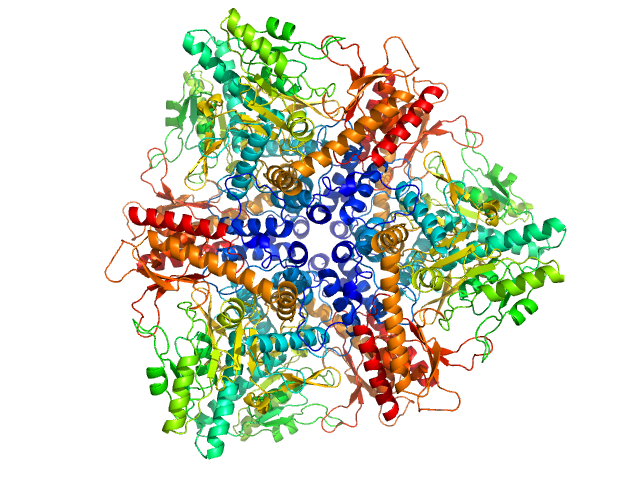
| Sample: |
Callose synthase octamer, 633 kDa Arabidopsis thaliana protein
|
| Buffer: |
Tris, 50 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Structural Characterization of Cell Wall and Plasma Membrane Proteins of Arabidopsis thaliana
University of Hamburg Dissertation 8022 (2016)
Haifa El Kilani
|
| RgGuinier |
8.0 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
1033 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glutamate decarboxylase alpha (GadA) from E. coli monomer, 53 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 29
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.4 |
nm |
| VolumePorod |
450 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
MHV-68 TR DNA monomer, 30 kDa unidentified herpesvirus DNA
|
| Buffer: |
10 mM TRIS 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
KLBS1-2 DNA monomer, 24 kDa unidentified herpesvirus DNA
|
| Buffer: |
Tris, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
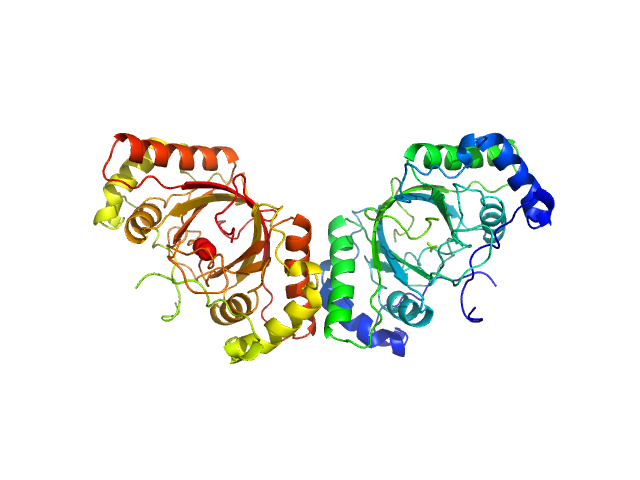
| Sample: |
ORF73 tetramer tetramer, 63 kDa Human herpesvirus 8 protein
ORF73 dimer dimer, 32 kDa Human herpesvirus 8 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
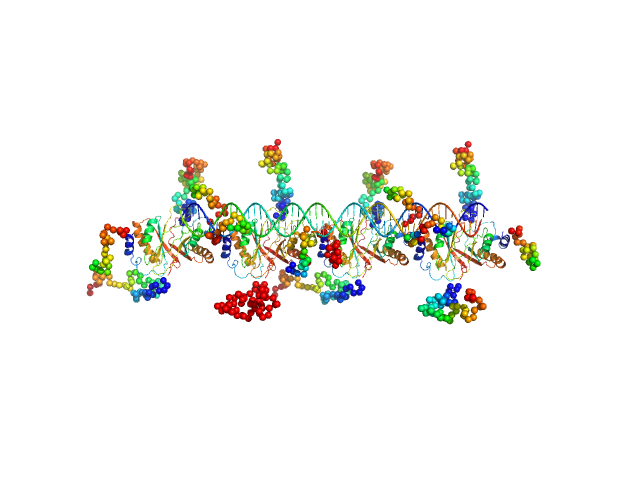
| Sample: |
MHV-68 TR DNA monomer, 30 kDa unidentified herpesvirus DNA
Latency-associated nuclear antigen octamer, 269 kDa Murid herpesvirus 4 protein
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
5.8 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
475 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
KLBS1-2 DNA monomer, 24 kDa unidentified herpesvirus DNA
ORF73 tetramer tetramer, 63 kDa Human herpesvirus 8 protein
ORF73 octamer octamer, 126 kDa Human herpesvirus 8 protein
KLBS1-2 DNA two monomers dimer, 48 kDa unidentified herpesvirus RNA
|
| Buffer: |
25 mM Na/K Phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Apr 27
|
KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA.
Nucleic Acids Res 43(20):10039-54 (2015)
Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
250 |
nm3 |
|
|