|
|
|
|
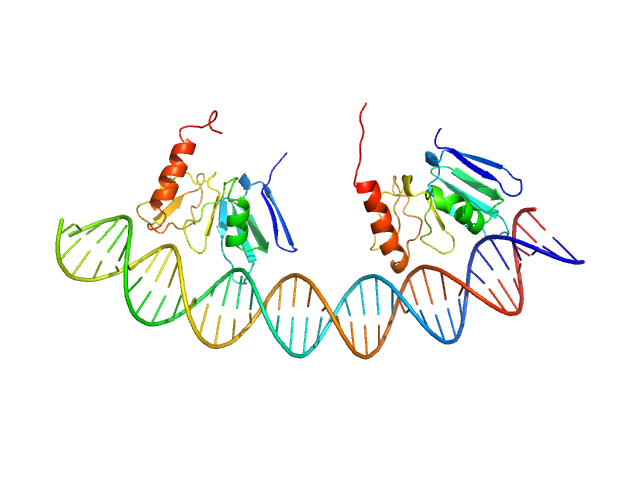
|
| Sample: |
Comcde, 24 kDa Streptococcus pneumoniae DNA
Response regulator dimer, 29 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM MES 500 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 6.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Jun 2
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
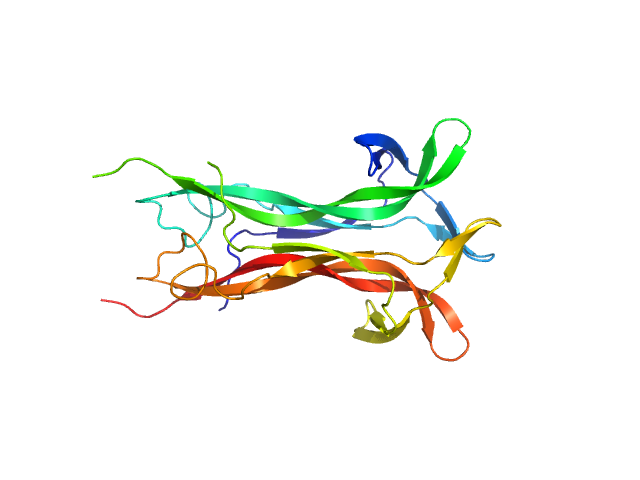
|
| Sample: |
Beta-nerve growth factor dimer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na-phosphate, 1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Jul 2
|
The conundrum of the high-affinity NGF binding site formation unveiled?
Biophys J 108(3):687-97 (2015)
Covaceuszach S, Konarev PV, Cassetta A, Paoletti F, Svergun DI, Lamba D, Cattaneo A
|
|
|
|
|
|
|
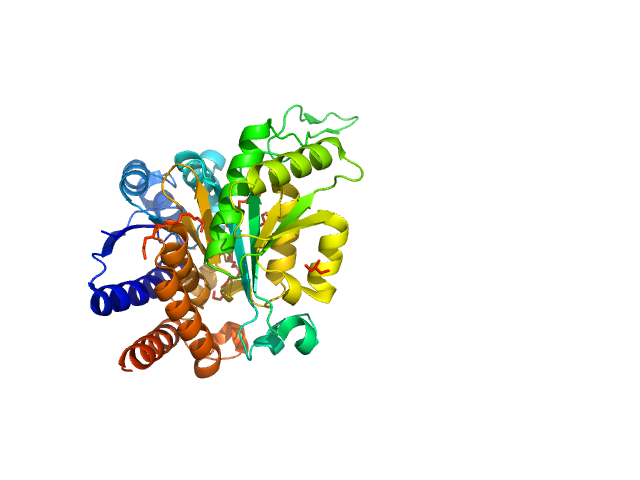
|
| Sample: |
3-isopropylmalate dehydrogenase dimer, 73 kDa Thermus thermophilus protein
|
| Buffer: |
25 mM MOPS/NaOH, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 3
|
Glutamate 270 plays an essential role in K(+)-activation and domain closure of Thermus thermophilus isopropylmalate dehydrogenase.
FEBS Lett 589(2):240-5 (2015)
Gráczer É, Palló A, Oláh J, Szimler T, Konarev PV, Svergun DI, Merli A, Závodszky P, Weiss MS, Vas M
|
|
|
|
|
|
|
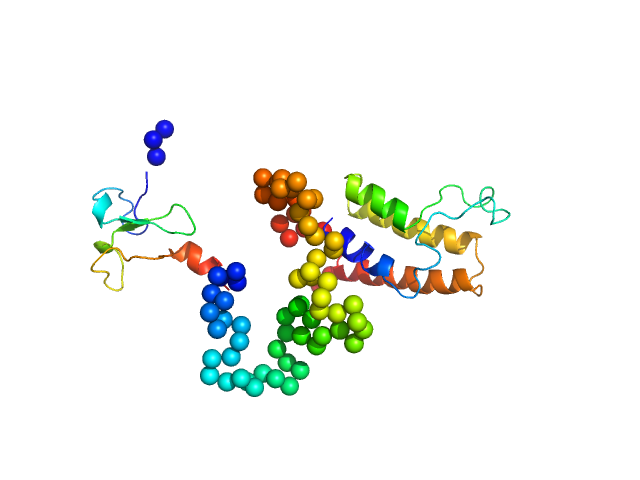
|
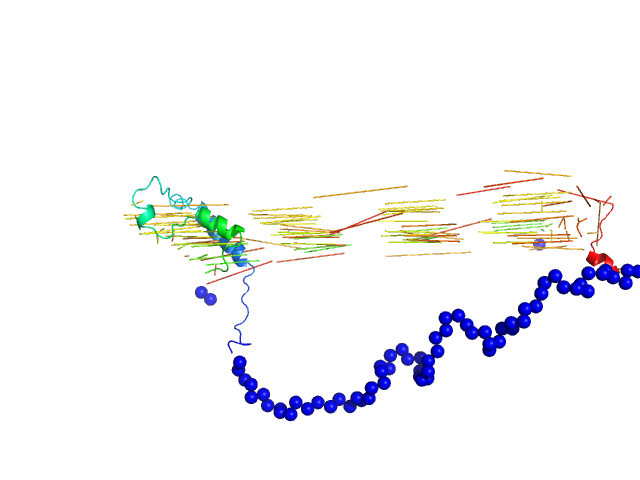
| Sample: |
Bromodomain adjacent to zinc finger domain protein 2A monomer, 27 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 500 mM NaCl 2 mM DTT 10 microM ZnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 16
|
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 23(1):80-92 (2015)
Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
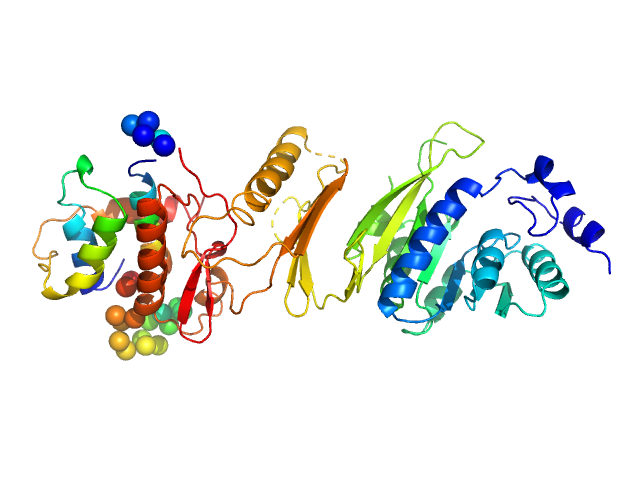
| Sample: |
Bromodomain adjacent to zinc finger domain protein 2B, C-terminal monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 500 mM NaCl 2 mM DTT 10 microM ZnCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 24
|
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 23(1):80-92 (2015)
Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
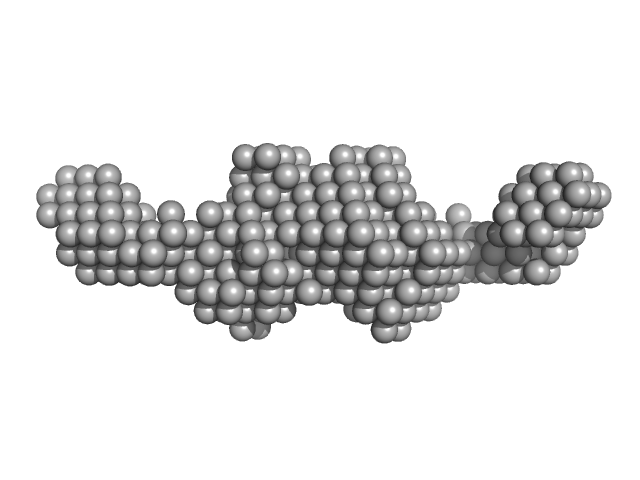
| Sample: |
Bifunctional kinase- methyltransferase WbdD monomer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM BisTris 50 mM NaCl 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Sep 23
|
A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide.
Nat Struct Mol Biol 22(1):50-56 (2015)
Hagelueken G, Clarke BR, Huang H, Tuukkanen A, Danciu I, Svergun DI, Hussain R, Liu H, Whitfield C, Naismith JH
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
|
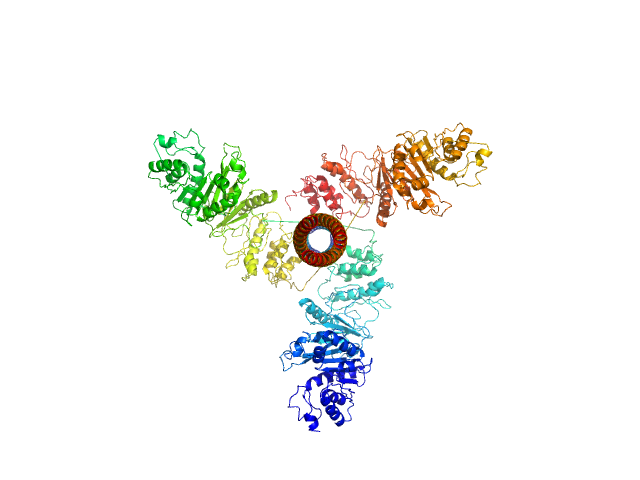
| Sample: |
Bifunctional kinase- methyltransferase WbdD trimer, 190 kDa protein
|
| Buffer: |
20 mM BisTris 50 mM NaCl 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 2
|
A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide.
Nat Struct Mol Biol 22(1):50-56 (2015)
Hagelueken G, Clarke BR, Huang H, Tuukkanen A, Danciu I, Svergun DI, Hussain R, Liu H, Whitfield C, Naismith JH
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RAID3 dimer, 23 kDa RNA
|
| Buffer: |
PBS 137 mM NaCl 2,7 mM KCl 6,5 mM Na2HPO4 1,5 mM KH2PO4, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2013 Oct 31
|
RAID3--An interleukin-6 receptor-binding aptamer with post-selective modification-resistant affinity.
RNA Biol 12(9):1043-53 (2015)
Mittelberger F, Meyer C, Waetzig GH, Zacharias M, Valentini E, Svergun DI, Berg K, Lorenzen I, Grötzinger J, Rose-John S, Hahn U
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Josephin domain of ataxin-3 monomer, 21 kDa Homo sapiens protein
Polyubiquitin-B monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 2 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 7
|
Allosteric regulation of deubiquitylase activity through ubiquitination.
Front Mol Biosci 2:2 (2015)
Faggiano S, Menon RP, Kelly GP, Todi SV, Scaglione KM, Konarev PV, Svergun DI, Paulson HL, Pastore A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
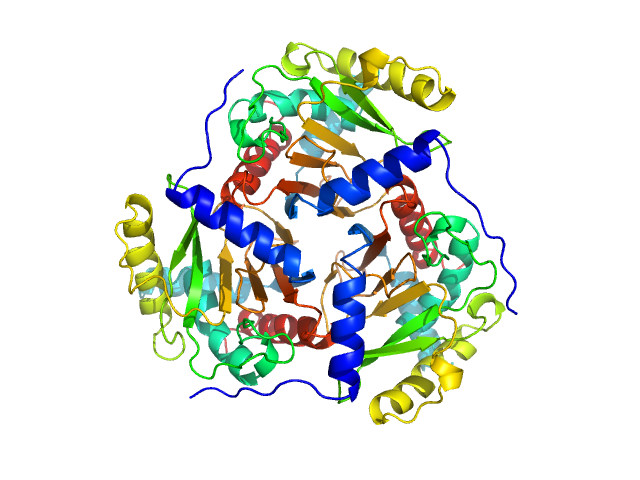
| Sample: |
Regulatory protein E2 trimer, 73 kDa Human papillomavirus type … protein
|
| Buffer: |
50 mM Tris ⁄ HCl, pH 8.8, 100 mM NaCl, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 27
|
Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex
Biochemical Journal 463(3):405-412 (2014)
Marrott N, Marshall J, Svergun D, Crennell S, Hough D, van den Elsen J, Danson M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
149 |
nm3 |
|
|