|
|
|
|
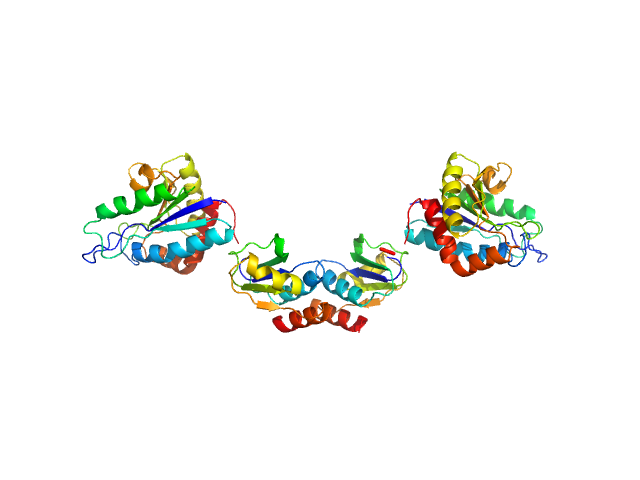
|
| Sample: |
Clostridium difficile bacteriophage 27 endolysin dimer, 64 kDa Clostridioides difficile protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 17
|
The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.
PLoS Pathog 10(7):e1004228 (2014)
Dunne M, Mertens HD, Garefalaki V, Jeffries CM, Thompson A, Lemke EA, Svergun DI, Mayer MJ, Narbad A, Meijers R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
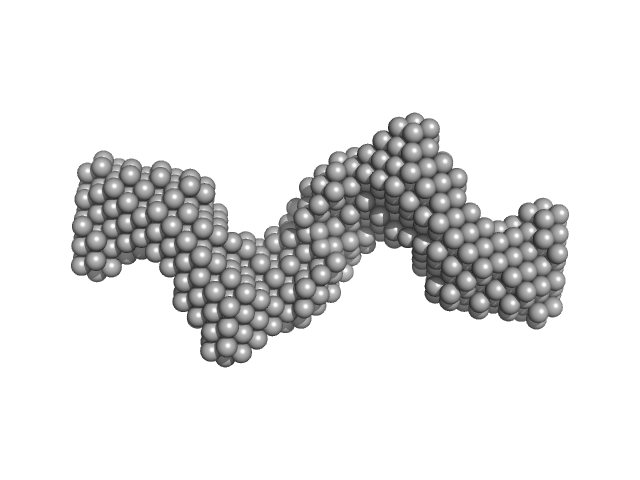
|
| Sample: |
Clostridium difficile bacteriophage 27 endolysin C238R mutant dimer, 64 kDa Clostridioides difficile protein
|
| Buffer: |
20 mM HEPES 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 21
|
The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.
PLoS Pathog 10(7):e1004228 (2014)
Dunne M, Mertens HD, Garefalaki V, Jeffries CM, Thompson A, Lemke EA, Svergun DI, Mayer MJ, Narbad A, Meijers R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
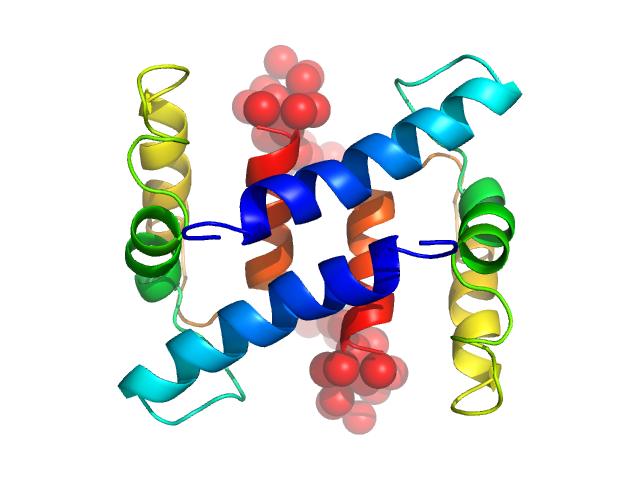
|
| Sample: |
Protein S100-A4 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 20 mM NaCl, 0.1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Mar 2
|
The C-Terminal Random Coil Region Tunes the Ca2+-Binding Affinity of S100A4 through Conformational Activation
PLoS ONE 9(5):e97654 (2014)
Duelli A, Kiss B, Lundholm I, Bodor A, Petoukhov M, Svergun D, Nyitray L, Katona G, Csernoch L
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
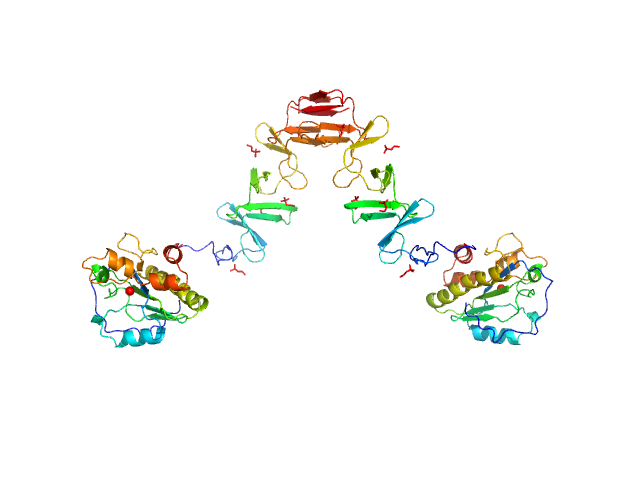
|
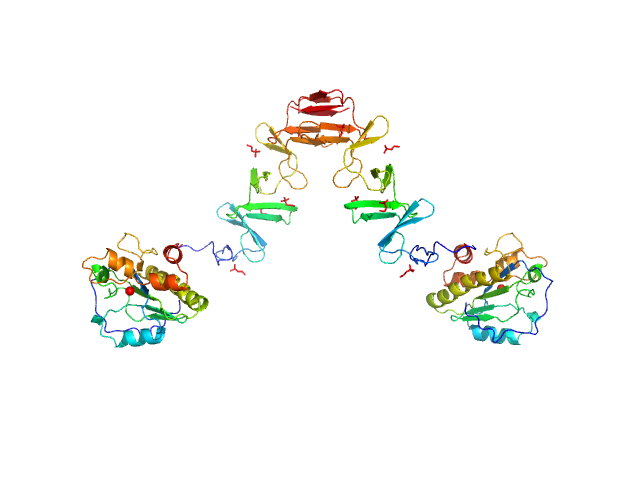
| Sample: |
Lytic Amidase with choline dimer, 75 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
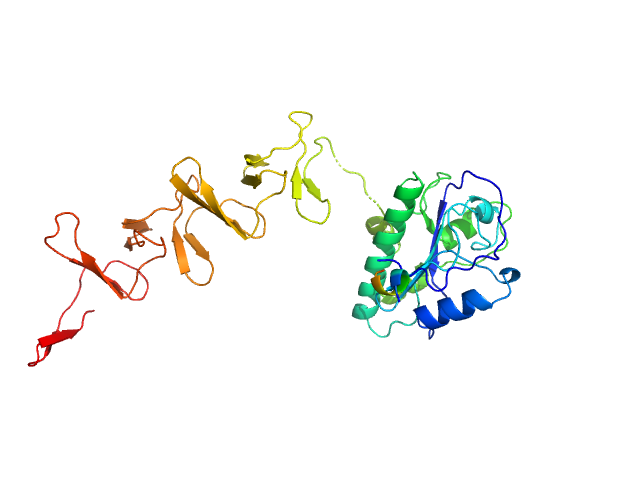
| Sample: |
Prophage Lytic Amidase with choline dimer, 73 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
5.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lytic Amidase without choline monomer, 37 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 3
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
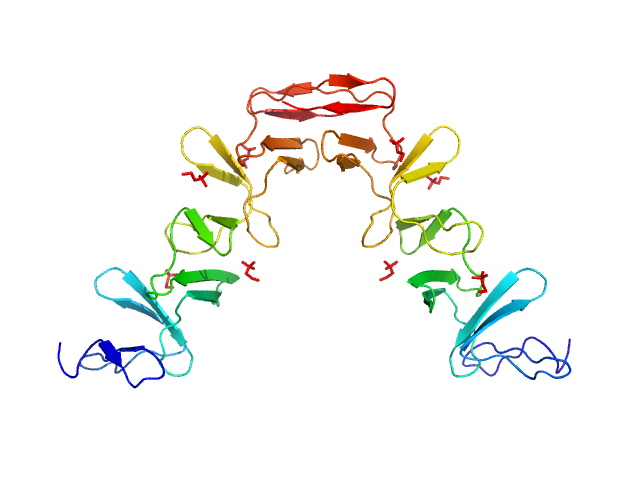
| Sample: |
Lytic Amidase choline-binding domain dimer, 17 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 5 mM choline chloride 1 µM ZnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 11
|
Structural and functional insights into peptidoglycan access for the lytic amidase LytA of Streptococcus pneumoniae.
MBio 5(1):e01120-13 (2014)
Mellroth P, Sandalova T, Kikhney A, Vilaplana F, Hesek D, Lee M, Mobashery S, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|

|
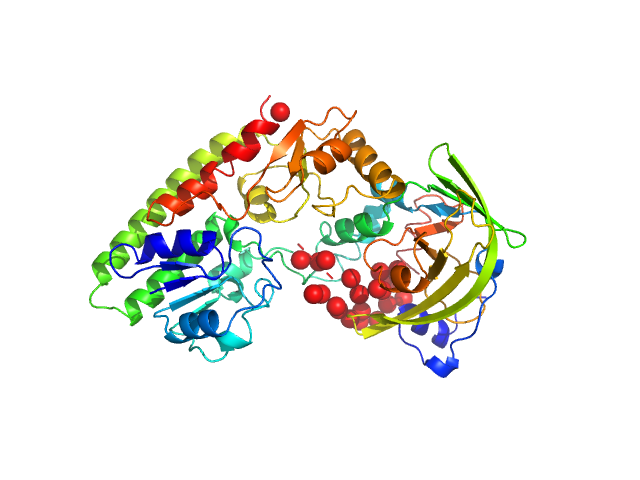
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2009 Nov 19
|
The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping.
Proc Natl Acad Sci U S A 111(1):457-62 (2014)
Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
High-affinity zinc transporter periplasmic component monomer, 33 kDa Salmonella enterica subsp. … protein
Zinc/cadmium-binding protein monomer, 23 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 May 9
|
The Salmonella enterica ZinT structure, zinc affinity and interaction with the high-affinity uptake protein ZnuA provide insight into the management of periplasmic zinc.
Biochim Biophys Acta 1840(1):535-44 (2014)
Ilari A, Alaleona F, Tria G, Petrarca P, Battistoni A, Zamparelli C, Verzili D, Falconi M, Chiancone E
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
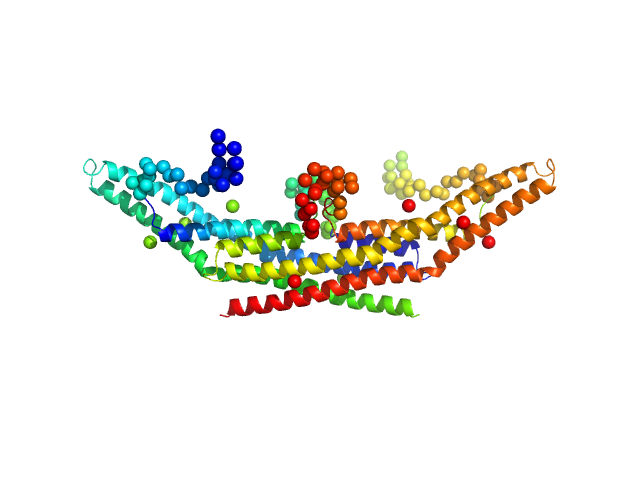
|
| Sample: |
Endophilin-A1 BAR domain dimer, 58 kDa Mus musculus protein
|
| Buffer: |
50 mM TRIS-HCL 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 13
|
Endophilin-A1 BAR domain interaction with arachidonyl CoA.
Front Mol Biosci 1:20 (2014)
Petoukhov MV, Weissenhorn W, Svergun DI
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|