|
|
|
|

|
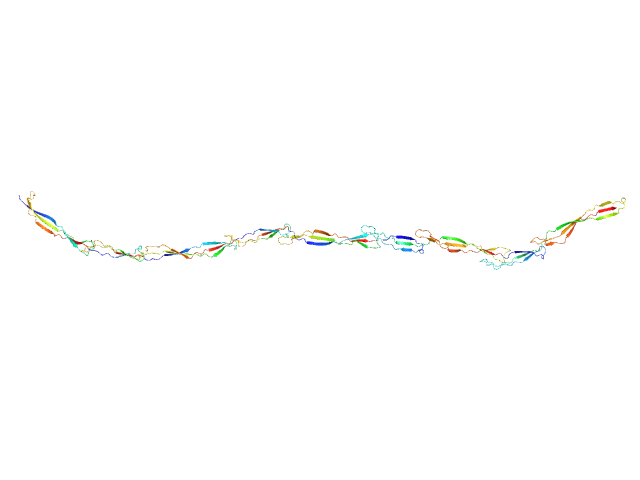
| Sample: |
Surface protein G monomer, 53 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
9.7 |
nm |
| Dmax |
38.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
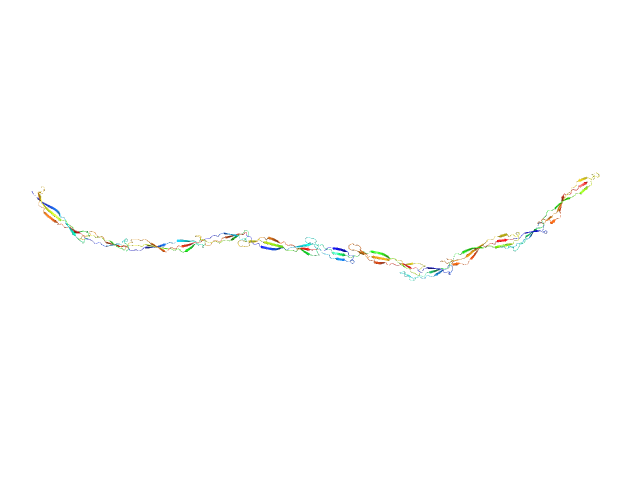
| Sample: |
Surface protein G monomer, 67 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
12.0 |
nm |
| Dmax |
48.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
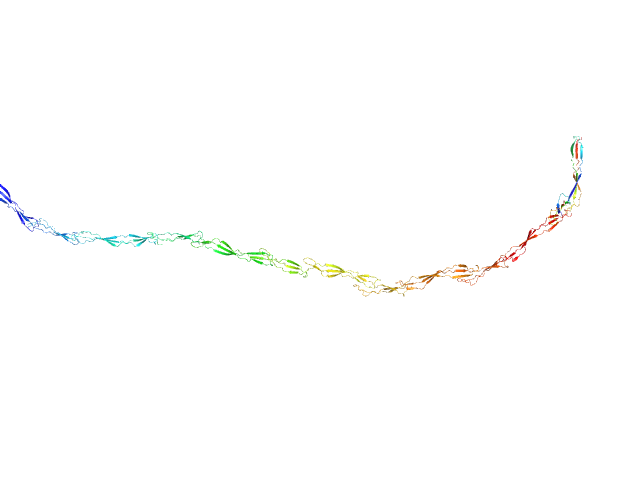
| Sample: |
Surface protein G monomer, 81 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
14.1 |
nm |
| Dmax |
57.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
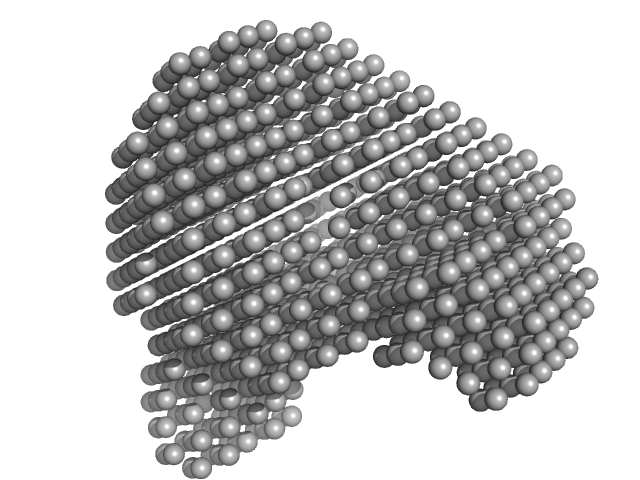
| Sample: |
Surface protein G monomer, 95 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
15.9 |
nm |
| Dmax |
63.0 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
20 mM Sodium Acetate/HEPES, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Feb 17
|
Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra.
Nat Methods 12(5):419-22 (2015)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human serum albumin monomer monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 22
|
Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra.
Nat Methods 12(5):419-22 (2015)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylose Isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
20 mM HEPES 200 mM Na2SO4 50 mM K2SO4 500 % v/v D2O 1 mM MgCl2, pH: 6.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 10
|
Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra.
Nat Methods 12(5):419-22 (2015)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
234 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Integrin beta-4 monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 6
|
Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin α6β4.
Acta Crystallogr D Biol Crystallogr 71(Pt 4):969-85 (2015)
Alonso-García N, García-Rubio I, Manso JA, Buey RM, Urien H, Sonnenberg A, Jeschke G, de Pereda JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
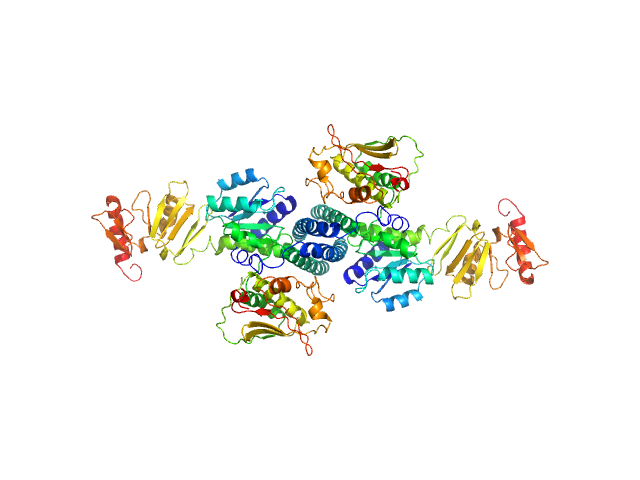
|
| Sample: |
Histidine protein kinase dimer, 54 kDa Streptococcus pneumoniae protein
Response regulator dimer, 61 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, IBBMC on 2012 May 16
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
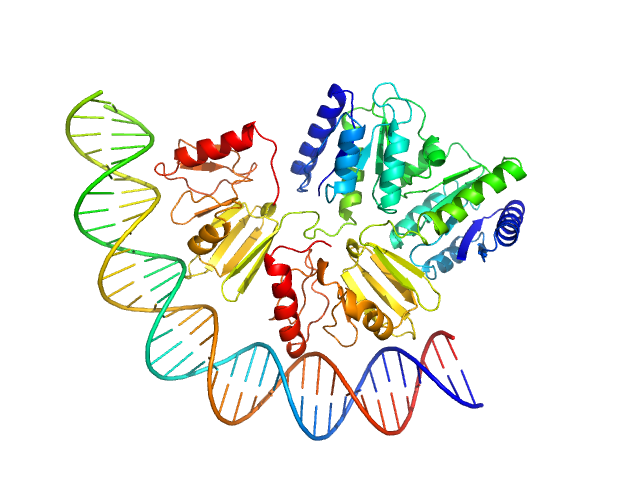
|
| Sample: |
Comcde, 24 kDa Streptococcus pneumoniae DNA
Response regulator dimer, 61 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM MES 500 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 6.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Feb 11
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
122 |
nm3 |
|
|