|
|
|
|
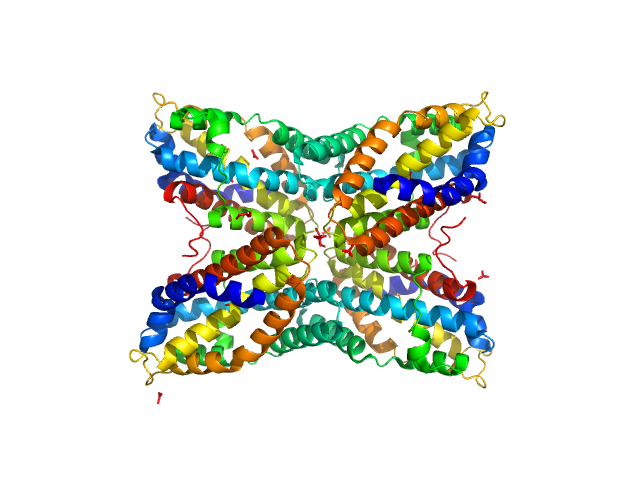
|
| Sample: |
Thiaminase type II enzyme tetramer, 107 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 11
|
Staphylococcus aureus thiaminase II: oligomerization warrants proteolytic protection against serine proteases.
Acta Crystallogr D Biol Crystallogr 69(Pt 12):2320-9 (2013)
Begum A, Drebes J, Kikhney A, Müller IB, Perbandt M, Svergun D, Wrenger C, Betzel C
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
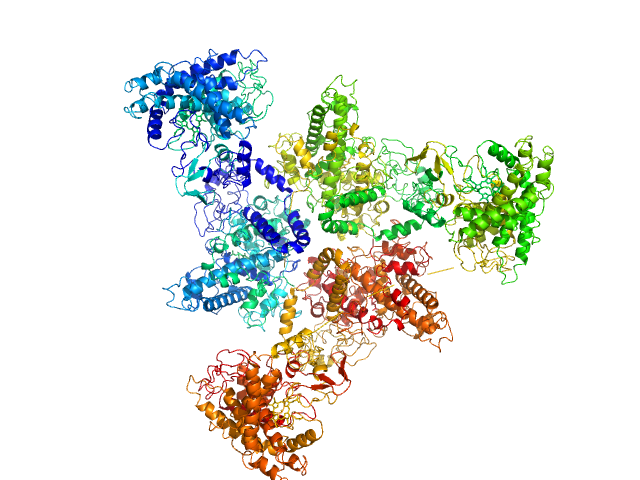
|
| Sample: |
Psi-producing oxygenase A trimer, 362 kDa Aspergillus nidulans protein
|
| Buffer: |
20 Mm HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 24
|
A structural model of PpoA derived from SAXS-analysis-implications for substrate conversion.
Biochim Biophys Acta 1831(9):1449-57 (2013)
Koch C, Tria G, Fielding AJ, Brodhun F, Valerius O, Feussner K, Braus GH, Svergun DI, Bennati M, Feussner I
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
540 |
nm3 |
|
|
|
|
|
|
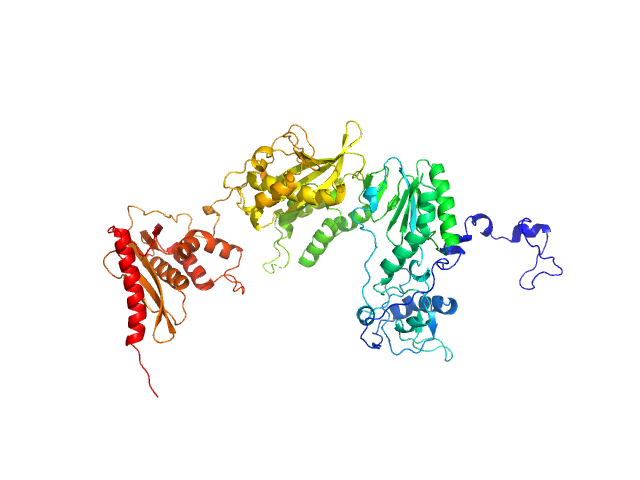
|
| Sample: |
Apo XMRV RT monomer, 75 kDa Escherichia coli protein
|
| Buffer: |
10 mM HEPES 100 mM KCl 5% Glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 8
|
Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid.
Nucleic Acids Res 41(6):3874-87 (2013)
Nowak E, Potrzebowski W, Konarev PV, Rausch JW, Bona MK, Svergun DI, Bujnicki JM, Le Grice SF, Nowotny M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
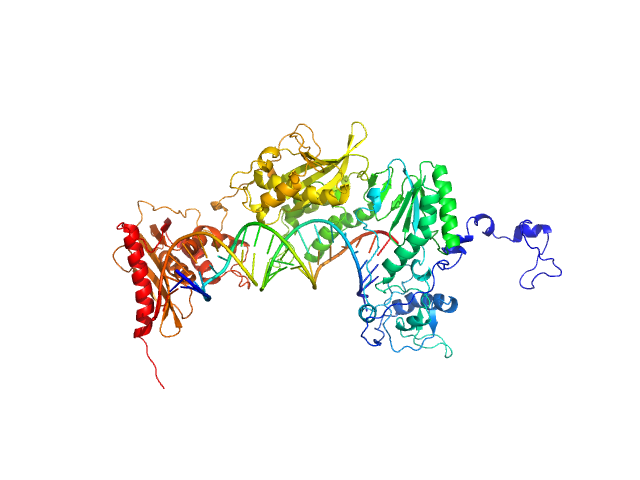
|
| Sample: |
Apo XMRV RT monomer, 75 kDa Escherichia coli protein
RNA_DNA hybrid substrate monomer, 15 kDa
|
| Buffer: |
10 mM HEPES 100 mM KCl 5% Glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 8
|
Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid.
Nucleic Acids Res 41(6):3874-87 (2013)
Nowak E, Potrzebowski W, Konarev PV, Rausch JW, Bona MK, Svergun DI, Bujnicki JM, Le Grice SF, Nowotny M
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|
|
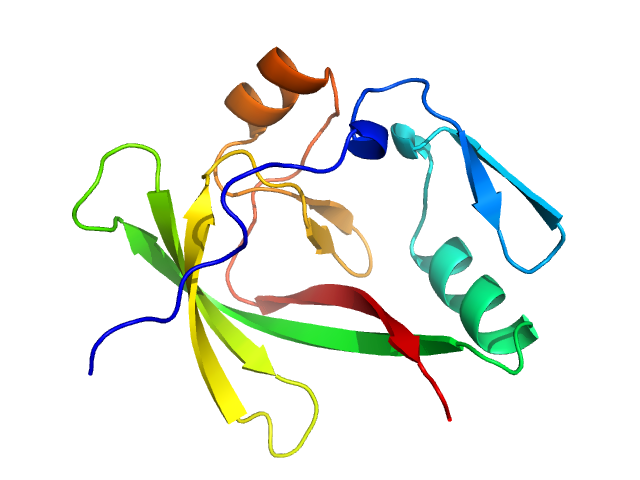
| Sample: |
Ataxin-1 monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 21
|
Self-assembly and conformational heterogeneity of the AXH domain of ataxin-1: an unusual example of a chameleon fold.
Biophys J 104(6):1304-13 (2013)
de Chiara C, Rees M, Menon RP, Pauwels K, Lawrence C, Konarev PV, Svergun DI, Martin SR, Chen YW, Pastore A
|
|
|
|
|
|
|
|
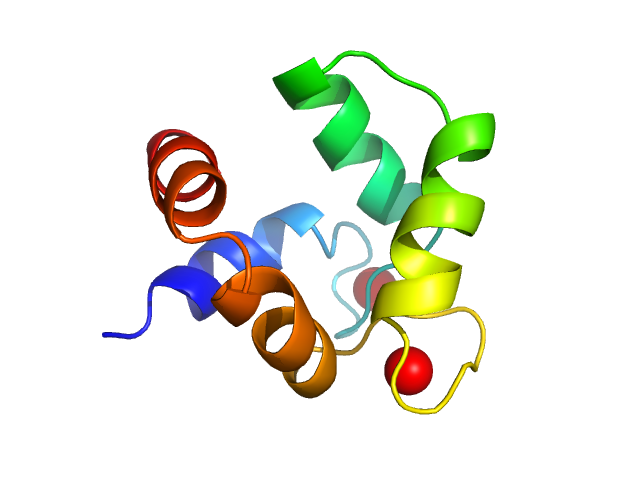
| Sample: |
Polcalcin Phl p 7 monomer, 9 kDa Phleum pratense protein
|
| Buffer: |
0.15M NaCl, 0.025M Hepes, pH 7.4, 100 uM Ca2+, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Dec 8
|
Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg2+-bound, and fully Ca2+-bound.
Proteins 81(2):300-15 (2013)
Henzl MT, Sirianni AG, Wycoff WG, Tan A, Tanner JJ
|
| RgGuinier |
1.3 |
nm |
| Dmax |
3.6 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
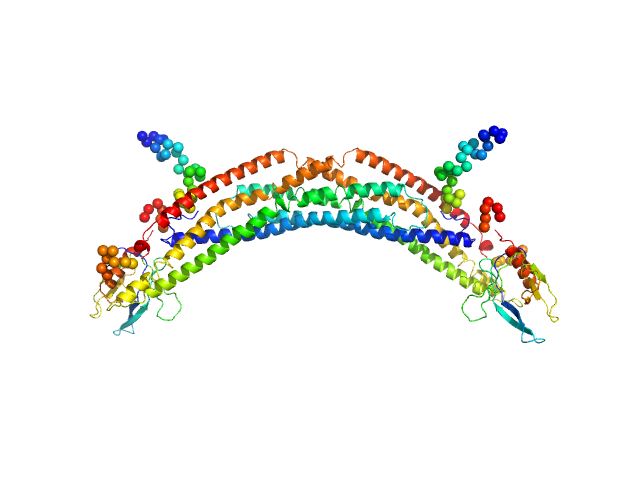
| Sample: |
Adaptor protein, phosphotyrosine interaction, pleckstrin homology domain, and leucine zipper-containing protein 2 dimer, 87 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 7 Apr 20
|
Membrane curvature protein exhibits interdomain flexibility and binds a small GTPase.
J Biol Chem 287(49):40996-1006 (2012)
King GJ, Stöckli J, Hu SH, Winnen B, Duprez WG, Meoli CC, Junutula JR, Jarrott RJ, James DE, Whitten AE, Martin JL
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
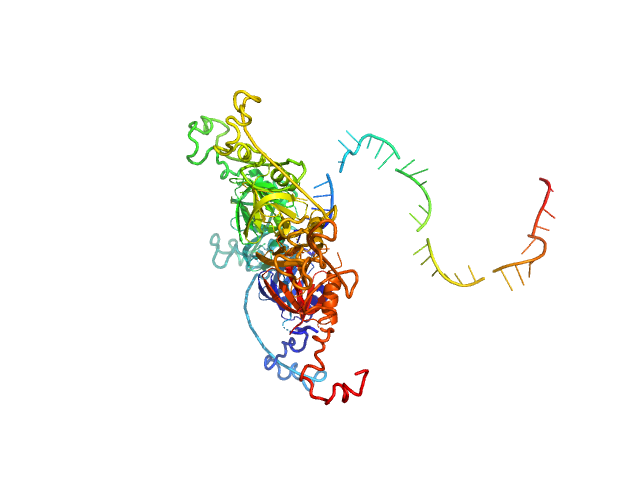
|
| Sample: |
RNA chaperone Hfq hexamer, 67 kDa Escherichia coli protein
RNA DsrA monomer, 12 kDa RNA
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Nov 16
|
Structural flexibility of RNA as molecular basis for Hfq chaperone function.
Nucleic Acids Res 40(16):8072-84 (2012)
Ribeiro Ede A Jr, Beich-Frandsen M, Konarev PV, Shang W, Vecerek B, Kontaxis G, Hämmerle H, Peterlik H, Svergun DI, Bläsi U, Djinović-Carugo K
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
210 |
nm3 |
|
|