|
|
|
|
|
| Sample: |
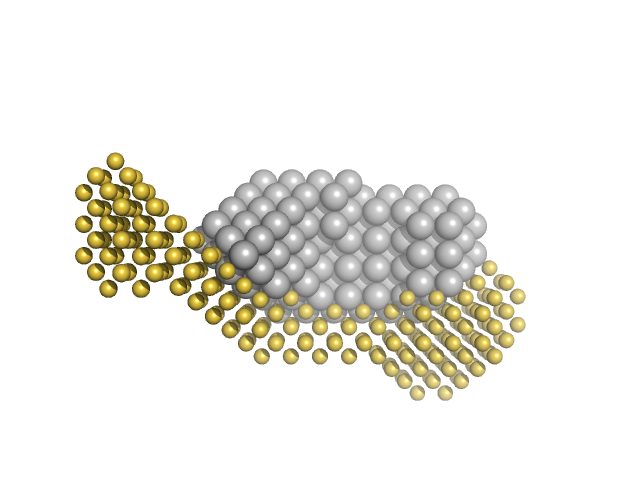
Calmodulin-1 monomer, 17 kDa Xenopus laevis protein
Calcium ions tetramer, 0 kDa
Gag-Pol polyprotein monomer, 15 kDa Human immunodeficiency virus … protein
|
| Buffer: |
50 mM MOPS, 5 mM CaCl2, 2 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, Australian Nuclear Science and Technology Organisation on 2010 Jun 23
|
Calmodulin binds a highly extended HIV-1 MA protein that refolds upon its release.
Biophys J 103(3):541-549 (2012)
Taylor JE, Chow JYH, Jeffries CM, Kwan AH, Duff AP, Hamilton WA, Trewhella J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
CYNEX4 FRET probe, (eYFP-AnnexinA4-eCFP) monomer, 93 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1 mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
CYNEX4 FRET probe, (eYFP-AnnexinA4-eCFP) T266D mutant monomer, 93 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|
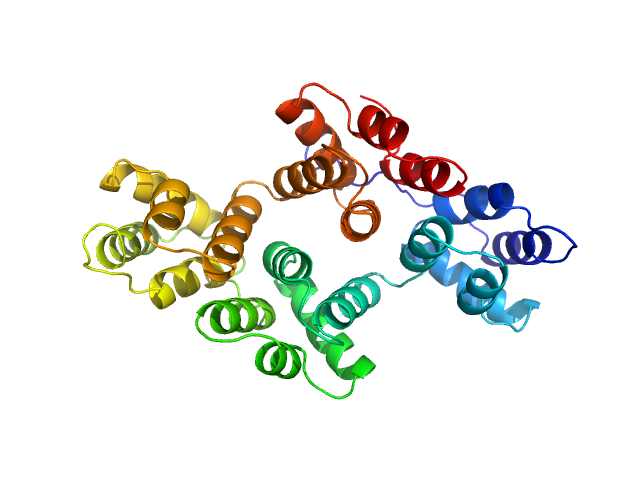
|
| Sample: |
Annexin-A4 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
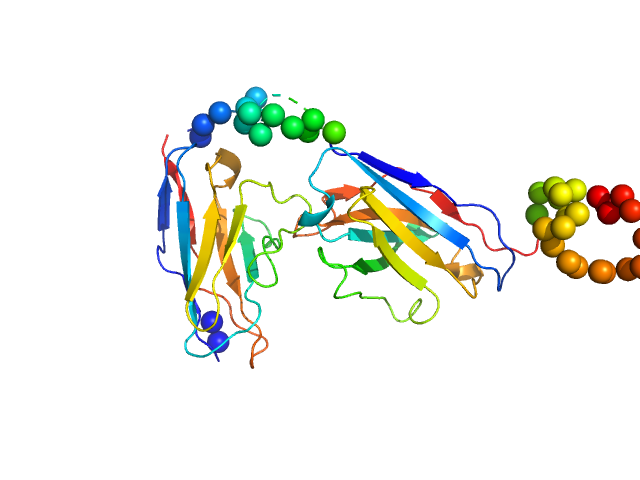
Recombinant monoclonal anti-proNGF antibody in single chain Fv fragment (scFv) monomer, 29 kDa protein
|
| Buffer: |
phosphate buffered saline, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 21
|
Direct intracellular selection and biochemical characterization of a recombinant anti-proNGF single chain antibody fragment.
Arch Biochem Biophys 522(1):26-36 (2012)
Paoletti F, Malerba F, Konarev PV, Visintin M, Scardigli R, Fasulo L, Lamba D, Svergun DI, Cattaneo A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
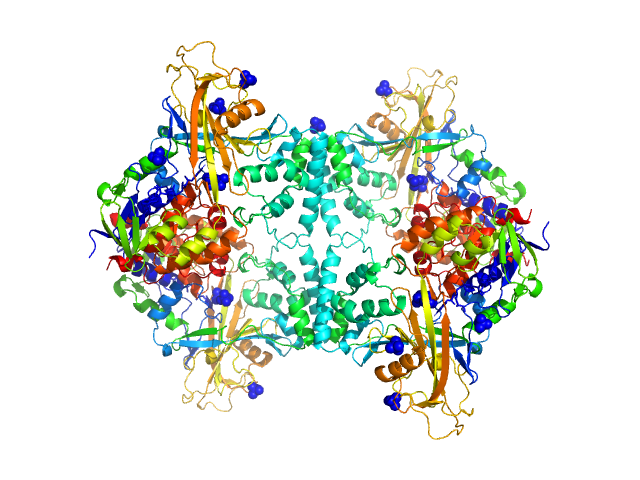
Aspergillus fumigatus UDP galactopyranose mutase tetramer, 228 kDa protein
|
| Buffer: |
20 mM HEPES, 45 mM NaCl, 0.5 mM Tris(hydroxypropyl)phosphine, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Apr 19
|
Crystal structures and small-angle x-ray scattering analysis of UDP-galactopyranose mutase from the pathogenic fungus Aspergillus fumigatus.
J Biol Chem 287(12):9041-51 (2012)
Dhatwalia R, Singh H, Oppenheimer M, Karr DB, Nix JC, Sobrado P, Tanner JJ
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
308 |
nm3 |
|
|
|
|
|
|
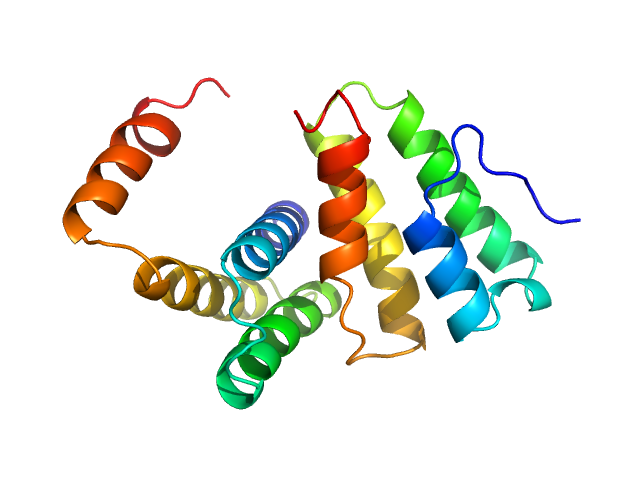
|
| Sample: |
Replicase polyprotein 1a dimer, 19 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Apr 2
|
Nonstructural Proteins 7 and 8 of Feline Coronavirus Form a 2:1 Heterotrimer That Exhibits Primer-Independent RNA Polymerase Activity
Journal of Virology 86(8):4444-4454 (2012)
Xiao Y, Ma Q, Restle T, Shang W, Svergun D, Ponnusamy R, Sczakiel G, Hilgenfeld R
|
|
|
|
|
|
|
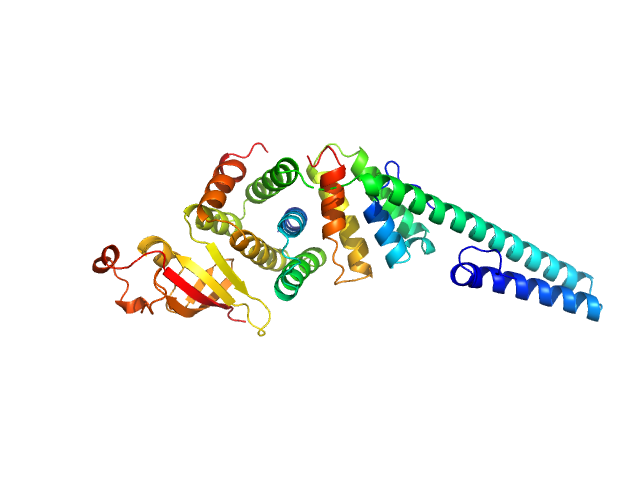
|
| Sample: |
Replicase polyprotein 1a dimer, 19 kDa Severe acute respiratory … protein
Replicase polyprotein 1a monomer, 22 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Apr 2
|
Nonstructural Proteins 7 and 8 of Feline Coronavirus Form a 2:1 Heterotrimer That Exhibits Primer-Independent RNA Polymerase Activity
Journal of Virology 86(8):4444-4454 (2012)
Xiao Y, Ma Q, Restle T, Shang W, Svergun D, Ponnusamy R, Sczakiel G, Hilgenfeld R
|
|
|
|
|
|
|
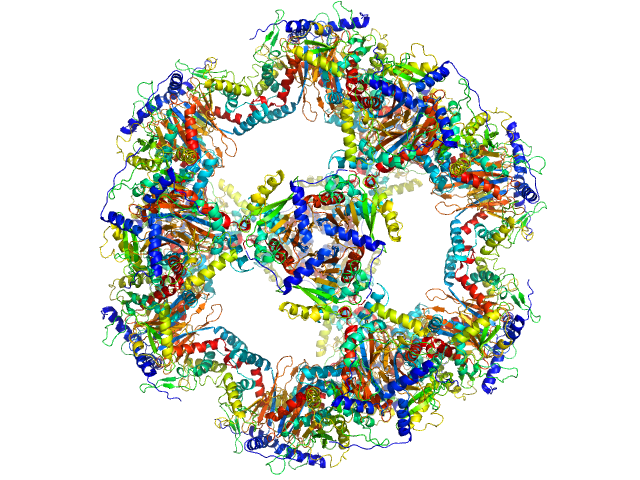
|
| Sample: |
Regulatory protein E2, 1037 kDa Human papillomavirus type … protein
|
| Buffer: |
50 mM Tris ⁄ HCl, pH 8.8, 100 mM NaCl, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Jul 13
|
The catalytic core of an archaeal 2-oxoacid dehydrogenase multienzyme complex is a 42-mer protein assembly
FEBS Journal 279(5):713-723 (2012)
Marrott N, Marshall J, Svergun D, Crennell S, Hough D, Danson M, van den Elsen J
|
| RgGuinier |
8.8 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
2473 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Full length GbpA monomer, 54 kDa Vibrio cholerae protein
|
| Buffer: |
25 mM Tris/HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 16
|
The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces.
PLoS Pathog 8(1):e1002373 (2012)
Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AF, Svergun DI, Eijsink VG, Chatterjee NS, van Aalten DM
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|