|
|
|
|
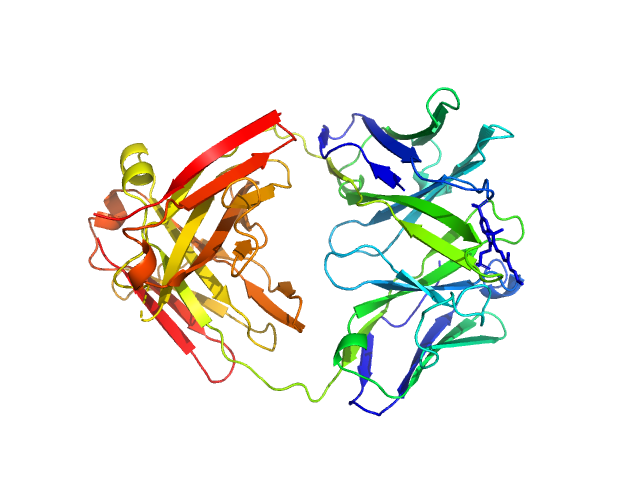
|
| Sample: |
Fab fragment in complex with small molecule hapten, crystal form-1 monomer, 45 kDa Homo sapiens protein
(1S)-1-AMINO-2-(1H-INDOL-3-YL)ETHANOL monomer, 0 kDa
|
| Buffer: |
20 mM Tris–HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 25
|
Fab MOR03268 Triggers Absorption Shift of a Diagnostic Dye via Packaging in a Solvent-shielded Fab Dimer Interface
Journal of Molecular Biology 377(1):206-219 (2008)
Hillig R, Urlinger S, Fanghänel J, Brocks B, Haenel C, Stark Y, Sülzle D, Svergun D, Baesler S, Malawski G, Moosmayer D, Menrad A, Schirner M, Licha K
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Calmodulin monomer, 17 kDa Homo sapiens protein
C-terminal region of human myelin basic protein monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris75 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 28
|
Interaction between the C-terminal region of human myelin basic protein and calmodulin: analysis of complex formation and solution structure.
BMC Struct Biol 8:10 (2008)
Majava V, Petoukhov MV, Hayashi N, Pirilä P, Svergun DI, Kursula P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
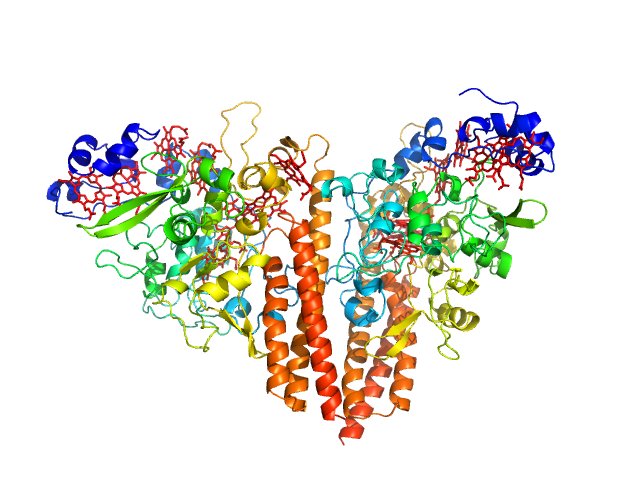
|
| Sample: |
Cytochrome c-552 hexamer, 356 kDa Thioalkalivibrio nitratireducens (strain … protein
|
| Buffer: |
Tris-borate buffer, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 8
|
Isolation and oligomeric composition of cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
Biochemistry (Mosc) 73(2):164-70 (2008)
Tikhonova TV, Slutskaya ES, Filimonenkov AA, Boyko KM, Kleimenov SY, Konarev PV, Polyakov KM, Svergun DI, Trofimov AA, Khomenkov VG, Zvyagilskaya RA, Popov VO
|
|
|
|
|
|
|
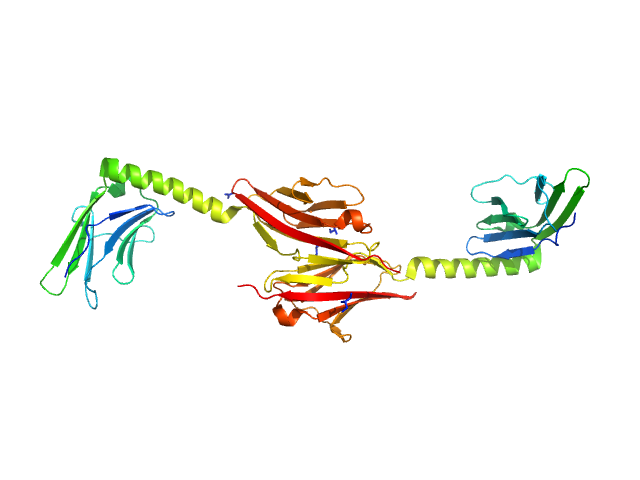
|
| Sample: |
Myomesin-1 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris/HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 7
|
Molecular basis of the C-terminal tail-to-tail assembly of the sarcomeric filament protein myomesin.
EMBO J 27(1):253-64 (2008)
Pinotsis N, Lange S, Perriard JC, Svergun DI, Wilmanns M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
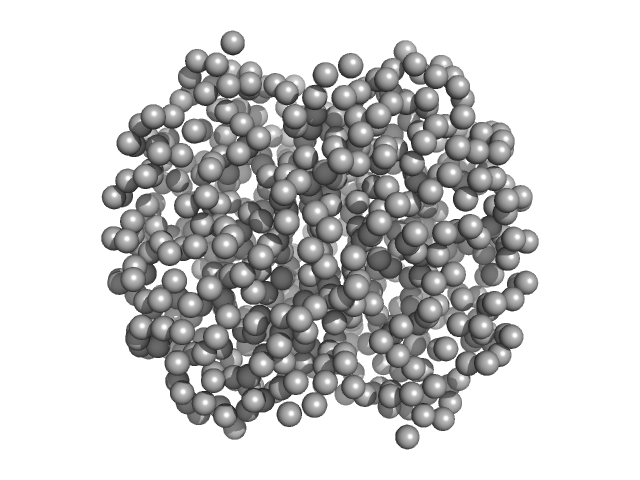
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
13.0 |
nm |
|
|
|
|
|
|
|
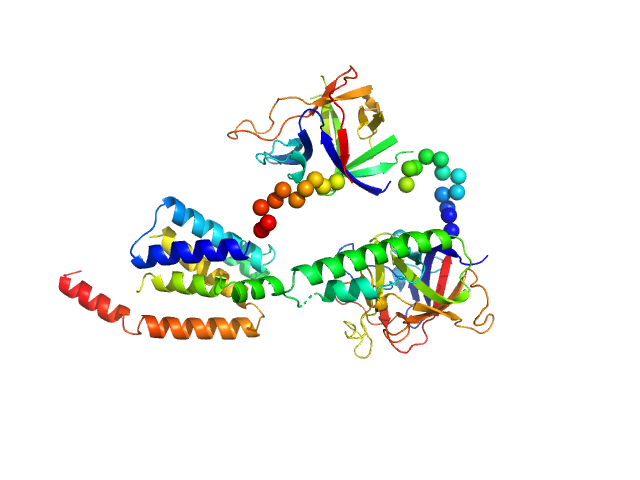
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 5 mM EGTA, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
|
|
|
|
|
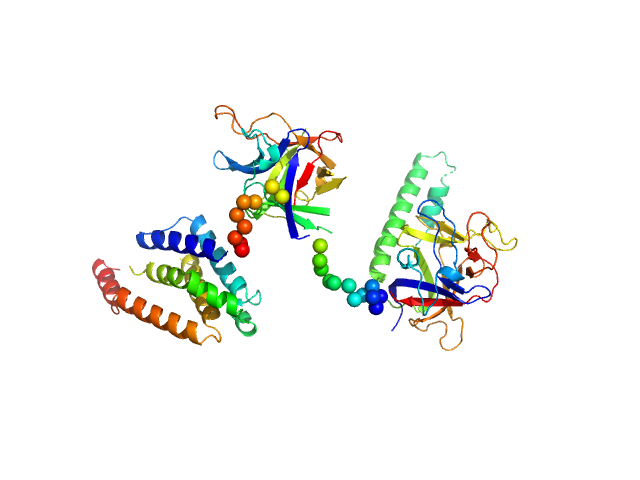
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
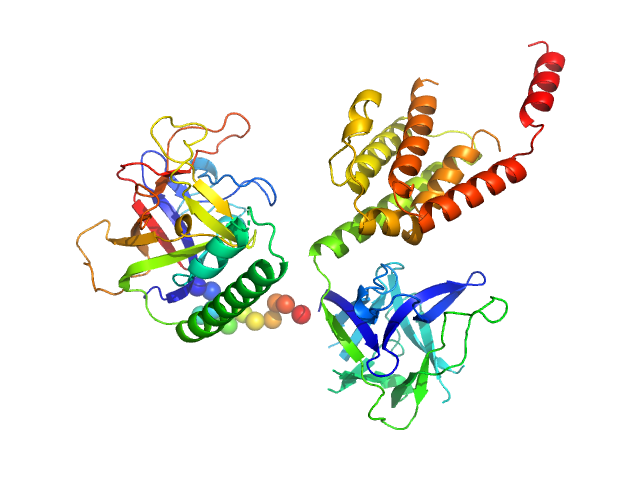
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 5 mM EGTA, 0.25 mM IP3, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
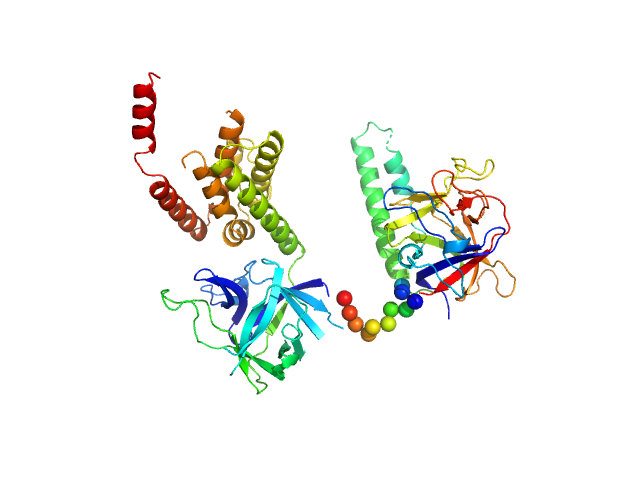
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 10 mM CaCl2, 0.25 mM IP3, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
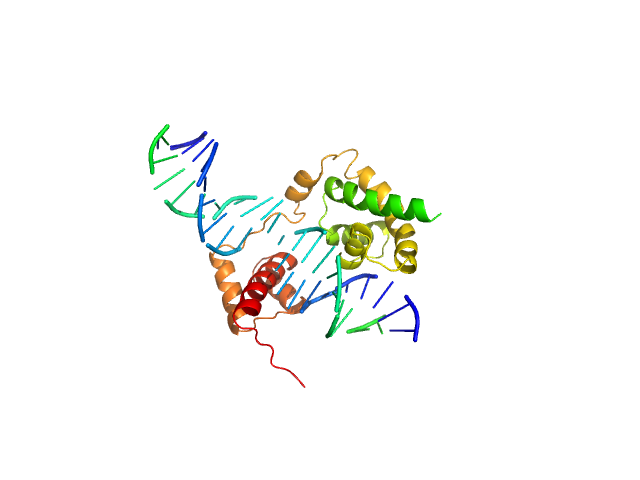
|
| Sample: |
POU domain, class 3, transcription factor 2 monomer, 19 kDa Homo sapiens protein
Rat CRH DNA monomer, 15 kDa Rattus norvegicus DNA
|
| Buffer: |
50 mM Tris, 0.4 M NaCl, 2% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2003 Oct 9
|
Fine-tuning of intrinsic N-Oct-3 POU domain allostery by regulatory DNA targets
Nucleic Acids Research 35(13):4420-4432 (2007)
Alazard R, Mourey L, Ebel C, Konarev P, Petoukhov M, Svergun D, Erard M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
41 |
nm3 |
|
|