|
|
|
|
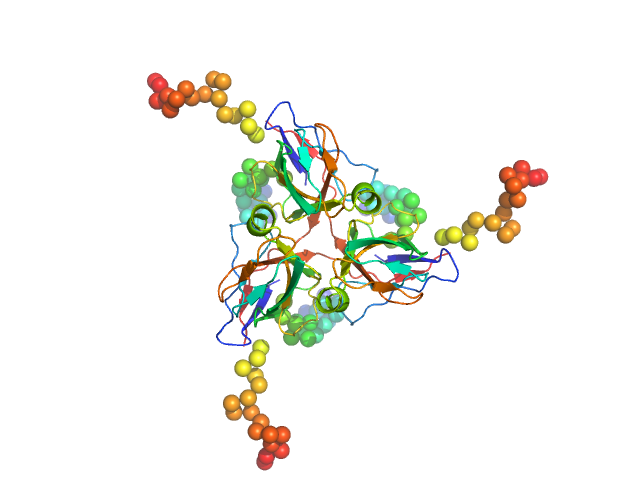
|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial trimer, 80 kDa Homo sapiens protein
|
| Buffer: |
20 mM citrate buffer, 1 mM DTT, 0.1 mM PMSF and 1 mM MgCl2, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Feb 1
|
Molecular shape and prominent role of β-strand swapping in organization of dUTPase oligomers
FEBS Letters 583(5):865-871 (2009)
Takács E, Barabás O, Petoukhov M, Svergun D, Vértessy B
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
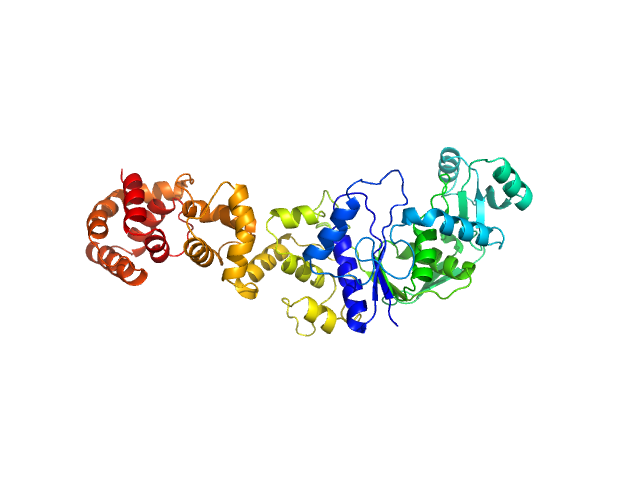
|
| Sample: |
Glutamate--tRNA ligase dimer, 108 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
35 mM HEPES⁄NaOH, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 19
|
Kinetic and mechanistic characterization of Mycobacterium tuberculosis glutamyl-tRNA synthetase and determination of its oligomeric structure in solution
FEBS Journal 276(5):1398-1417 (2009)
Paravisi S, Fumagalli G, Riva M, Morandi P, Morosi R, Konarev P, Petoukhov M, Bernier S, Chênevert R, Svergun D, Curti B, Vanoni M
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Der p21, allegren dimer, 26 kDa Escherichia coli protein
|
| Buffer: |
10 mM Hepes , pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 24
|
Characterization of Der p 21, a new important allergen derived from the gut of house dust mites.
Allergy 63(6):758-67 (2008)
Weghofer M, Dall'Antonia Y, Grote M, Stöcklinger A, Kneidinger M, Balic N, Krauth MT, Fernández-Caldas E, Thomas WR, van Hage M, Vieths S, Spitzauer S, Horak F, Svergun DI, Konarev PV, Valent P, Thalhamer J, Keller W, Valenta R, Vrtala S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytochrome C monomer, 11 kDa Escherichia coli protein
Adrenodoxin monomer, 11 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES 2 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jun 15
|
Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy.
J Am Chem Soc 130(20):6395-403 (2008)
Xu X, Reinle W, Hannemann F, Konarev PV, Svergun DI, Bernhardt R, Ubbink M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytochrome C dimer dimer, 22 kDa Escherichia coli protein
Adrenodoxin dimer dimer, 22 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES 2 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jun 15
|
Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy.
J Am Chem Soc 130(20):6395-403 (2008)
Xu X, Reinle W, Hannemann F, Konarev PV, Svergun DI, Bernhardt R, Ubbink M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
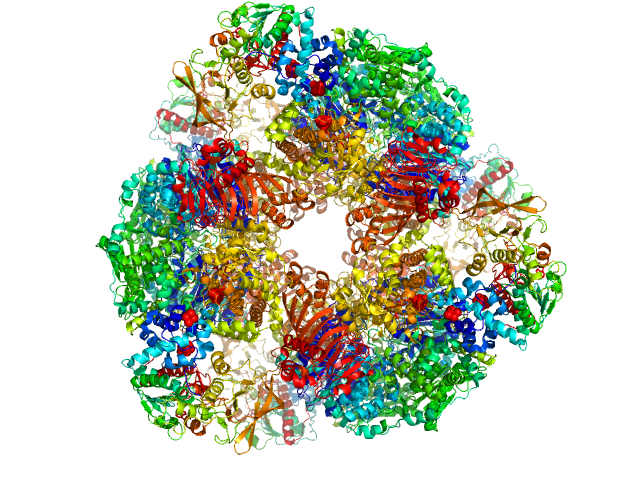
|
| Sample: |
Glutamate synthase [NADPH] large chain hexamer, 968 kDa Azospirillum brasilense protein
Glutamate synthase [NADPH] small chain hexamer, 296 kDa Azospirillum brasilense protein
|
| Buffer: |
25 mM Hepes/KOH, 1 mM EDTA, 1 mM dithiothreitol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 19
|
The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-MDa Hexamer by Cryoelectron Microscopy and Its Oligomerization Behavior in Solution
Journal of Biological Chemistry 283(13):8237-8249 (2008)
Cottevieille M, Larquet E, Jonic S, Petoukhov M, Caprini G, Paravisi S, Svergun D, Vanoni M, Boisset N
|
| RgGuinier |
7.7 |
nm |
| Dmax |
22.4 |
nm |
|
|
|
|
|
|
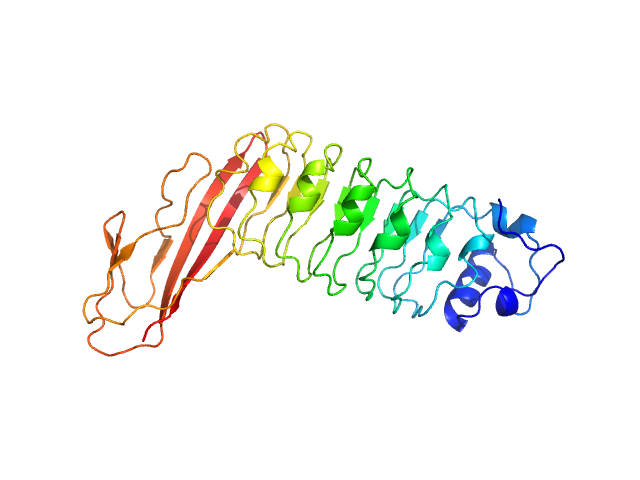
|
| Sample: |
Internalin B monomer, 36 kDa Listeria monocytogenes serotype … protein
|
| Buffer: |
25mM Na-phosphate buffer, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2003 Nov 11
|
X-ray and Neutron Small-Angle Scattering Analysis of the Complex Formed by the Met Receptor and the Listeria monocytogenes Invasion Protein InlB
Journal of Molecular Biology 377(2):489-500 (2008)
Niemann H, Petoukhov M, Härtlein M, Moulin M, Gherardi E, Timmins P, Heinz D, Svergun D
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
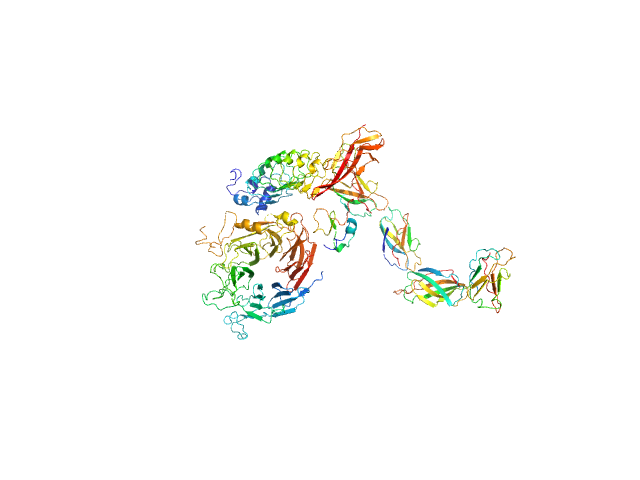
|
| Sample: |
Hepatocyte growth factor receptor monomer, 100 kDa Mus musculus protein
Internalin B monomer, 36 kDa Listeria monocytogenes serotype … protein
|
| Buffer: |
25mM Na-phosphate buffer, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Mar 1
|
X-ray and Neutron Small-Angle Scattering Analysis of the Complex Formed by the Met Receptor and the Listeria monocytogenes Invasion Protein InlB
Journal of Molecular Biology 377(2):489-500 (2008)
Niemann H, Petoukhov M, Härtlein M, Moulin M, Gherardi E, Timmins P, Heinz D, Svergun D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fab fragment in complex with small molecule hapten, crystal form-1 monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris–HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 25
|
Fab MOR03268 Triggers Absorption Shift of a Diagnostic Dye via Packaging in a Solvent-shielded Fab Dimer Interface
Journal of Molecular Biology 377(1):206-219 (2008)
Hillig R, Urlinger S, Fanghänel J, Brocks B, Haenel C, Stark Y, Sülzle D, Svergun D, Baesler S, Malawski G, Moosmayer D, Menrad A, Schirner M, Licha K
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fab fragment in complex with small molecule hapten, crystal form-1 monomer, 45 kDa Homo sapiens protein
(1S)-1-AMINO-2-(1H-INDOL-3-YL)ETHANOL monomer, 0 kDa
|
| Buffer: |
20 mM Tris–HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 27
|
Fab MOR03268 Triggers Absorption Shift of a Diagnostic Dye via Packaging in a Solvent-shielded Fab Dimer Interface
Journal of Molecular Biology 377(1):206-219 (2008)
Hillig R, Urlinger S, Fanghänel J, Brocks B, Haenel C, Stark Y, Sülzle D, Svergun D, Baesler S, Malawski G, Moosmayer D, Menrad A, Schirner M, Licha K
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
139 |
nm3 |
|
|