|
|
|
|
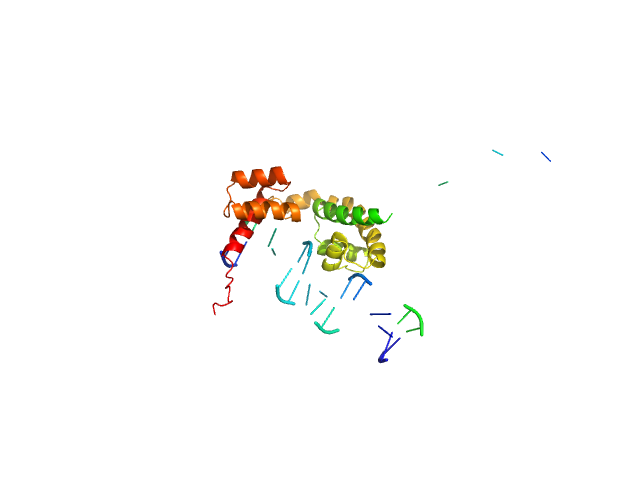
|
| Sample: |
POU domain, class 3, transcription factor 2 monomer, 19 kDa Homo sapiens protein
Human DR-alpha DNA monomer, 15 kDa Homo sapiens DNA
|
| Buffer: |
50 mM Tris, 0.4 M NaCl, 2% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 May 26
|
Fine-tuning of intrinsic N-Oct-3 POU domain allostery by regulatory DNA targets
Nucleic Acids Research 35(13):4420-4432 (2007)
Alazard R, Mourey L, Ebel C, Konarev P, Petoukhov M, Svergun D, Erard M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Filamin C 23-24 dimer, 40 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jun 17
|
Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23-24 dimer.
J Mol Biol 368(4):1011-23 (2007)
Sjekloća L, Pudas R, Sjöblom B, Konarev P, Carugo O, Rybin V, Kiema TR, Svergun D, Ylänne J, Djinović Carugo K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
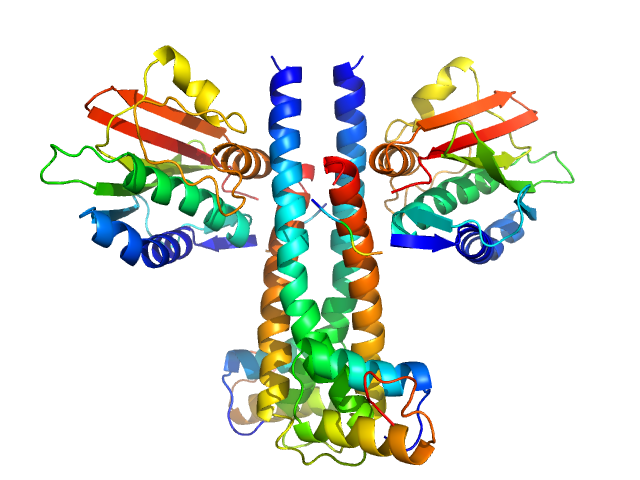
|
| Sample: |
Sporulation kinase A dimer, 51 kDa Bacillus subtilis protein
Sporulation inhibitor sda dimer, 11 kDa Bacillus subtilis protein
|
| Buffer: |
50mM Tris, 200mM NaCl, 150mM Imidazole, pH: 8.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Nov 16
|
The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition.
J Mol Biol 368(2):407-20 (2007)
Whitten AE, Jacques DA, Hammouda B, Hanley T, King GF, Guss JM, Trewhella J, Langley DB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
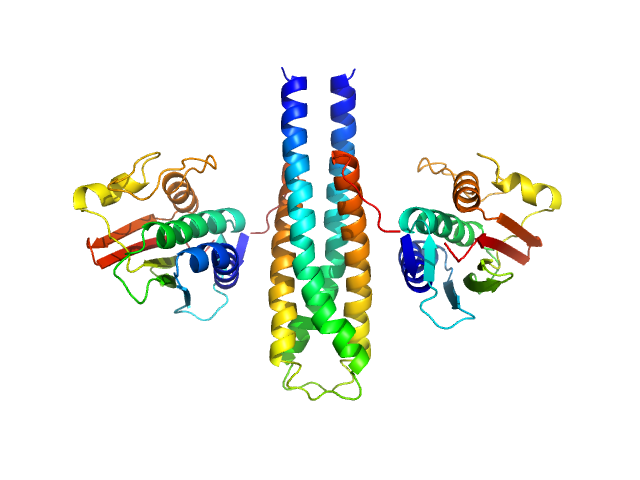
|
| Sample: |
Sporulation kinase A dimer, 51 kDa Bacillus subtilis protein
|
| Buffer: |
50mM Tris, 200mM NaCl, 150mM Imidazole, pH: 8.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Mar 18
|
The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition.
J Mol Biol 368(2):407-20 (2007)
Whitten AE, Jacques DA, Hammouda B, Hanley T, King GF, Guss JM, Trewhella J, Langley DB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
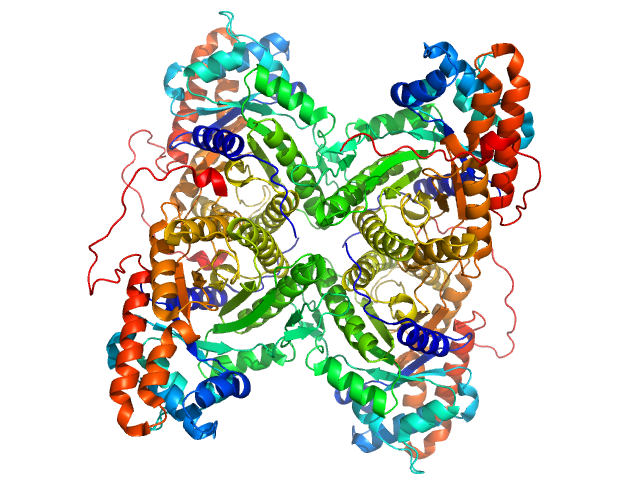
|
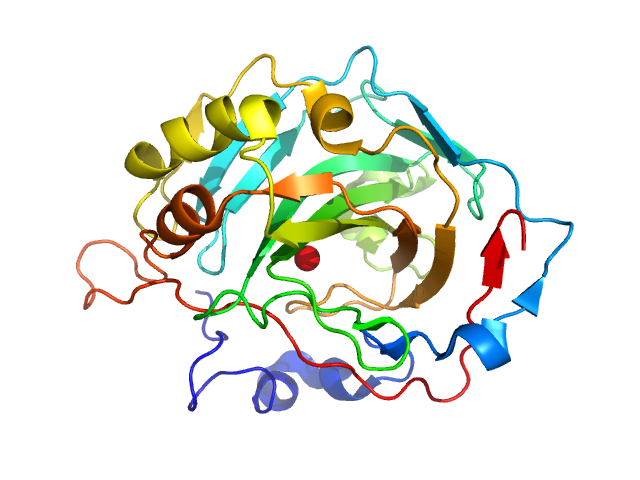
| Sample: |
Fructose-bisphosphate aldolase A tetramer, 157 kDa Oryctolagus cuniculus protein
|
| Buffer: |
100 mM Tris/HCl 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 19
|
Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering
Journal of Applied Crystallography 40(s1):s245-s249 (2007)
Mylonas E, Svergun D
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.5 |
nm |
|
|
|
|
|
|
|
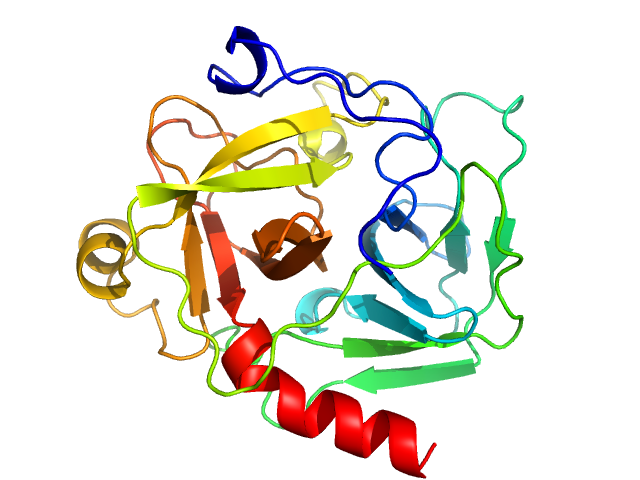
| Sample: |
Carbonic anhydrase 2 monomer, 29 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris/HCl 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 19
|
Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering
Journal of Applied Crystallography 40(s1):s245-s249 (2007)
Mylonas E, Svergun D
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.0 |
nm |
|
|
|
|
|
|
|
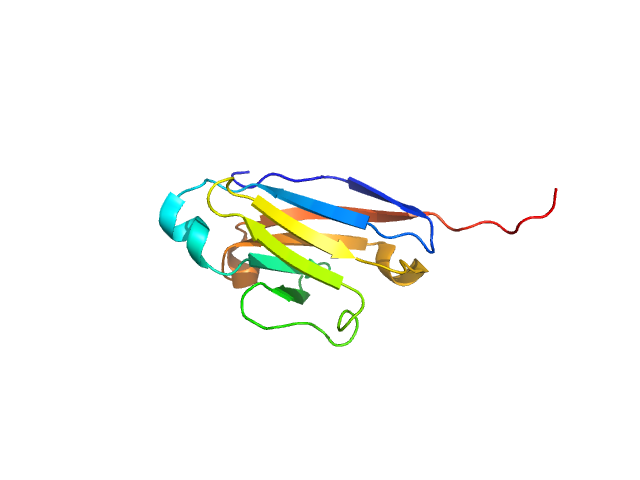
| Sample: |
Chymotrypsinogen A monomer, 26 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris/HCl 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 19
|
Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering
Journal of Applied Crystallography 40(s1):s245-s249 (2007)
Mylonas E, Svergun D
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.0 |
nm |
|
|
|
|
|
|
|
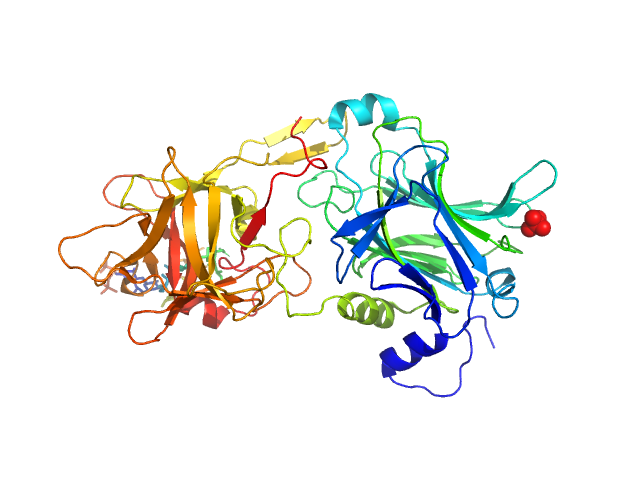
| Sample: |
CMRF35-like molecule 8 monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
MES buffer with 3mM DTT, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Apr 6
|
Molecular analysis and solution structure from small-angle X-ray scattering of the human natural killer inhibitory receptor IRp60 (CD300a)
International Journal of Biological Macromolecules 40(3):193-200 (2007)
Dimasi N, Roessle M, Moran O, Candiano G, Svergun D, Biassoni R
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Hc fragment of Tetanus toxin dimer, 103 kDa Clostridium tetani (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 300 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 May 14
|
The HC fragment of tetanus toxin forms stable, concentration-dependent dimers via an intermolecular disulphide bond.
J Mol Biol 365(1):123-34 (2007)
Qazi O, Bolgiano B, Crane D, Svergun DI, Konarev PV, Yao ZP, Robinson CV, Brown KA, Fairweather N
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
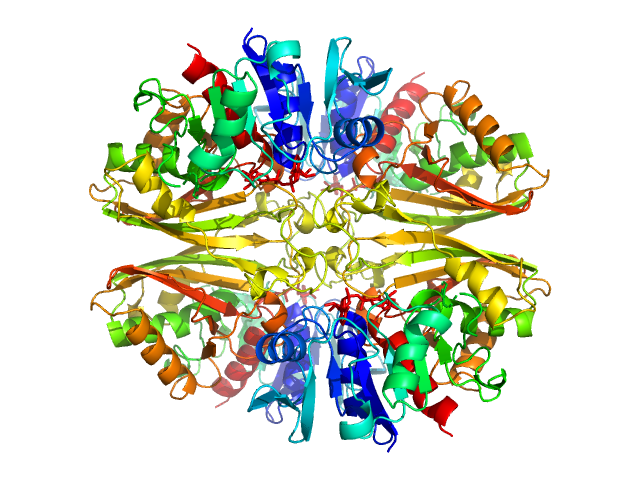
|
| Sample: |
Glyceraldehyde-3-phosphate dehydrogenase 1 tetramer, 142 kDa Kluyveromyces marxianus protein
|
| Buffer: |
150 mM NaCl, 1 mM beta-mercaptoethanol, 1 mM EDTA, 10 mM TrisHCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Mar 27
|
The Crystal and Solution Structures of Glyceraldehyde-3-phosphate Dehydrogenase Reveal Different Quaternary Structures
Journal of Biological Chemistry 281(44):33433-33440 (2006)
Ferreira-da-Silva F, Pereira P, Gales L, Roessle M, Svergun D, Moradas-Ferreira P, Damas A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
234 |
nm3 |
|
|