|
|
|
|

|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
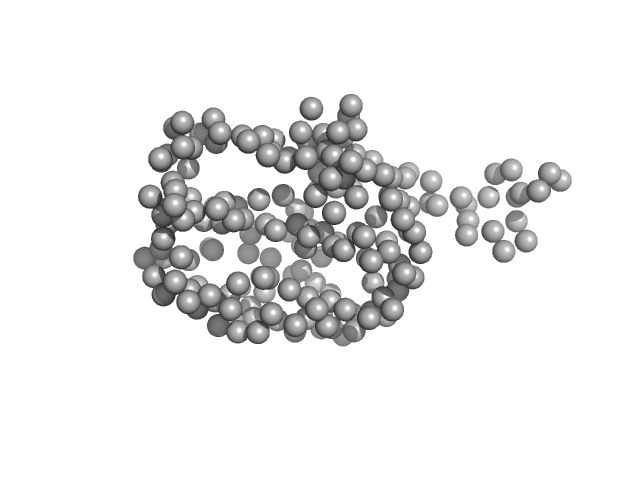
|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion) 24-mer, 755 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Jul 30
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
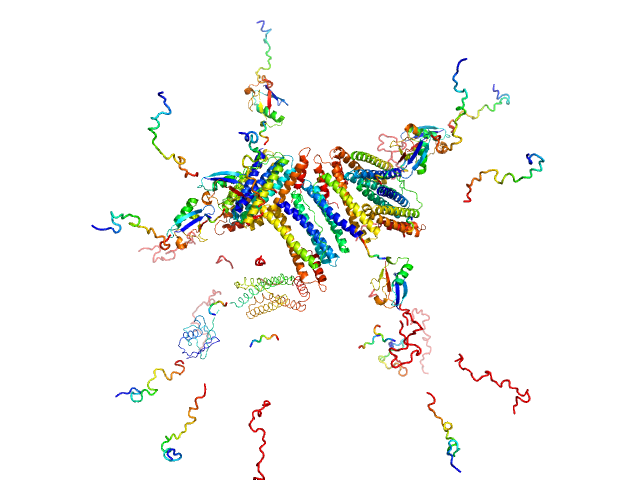
| RgGuinier |
7.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
2010 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion) octamer, 252 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Jul 17
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
618 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V) 24-mer, 480 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Dec 14
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
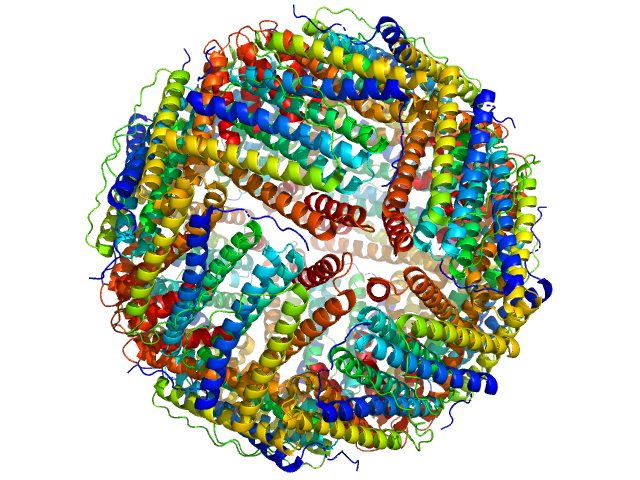
| RgGuinier |
5.6 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
674 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Heme acquisition protein HasAp dimer, 38 kDa Pseudomonas fluorescens (strain … protein
|
| Buffer: |
50 mM CHES, 5 % glycerol, pH: 9.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2023 Feb 24
|
Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom
RSC Advances 14(13):8829-8836 (2024)
Inaba H, Shisaka Y, Ariyasu S, Sakakibara E, Ueda G, Aiba Y, Shimizu N, Sugimoto H, Shoji O
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
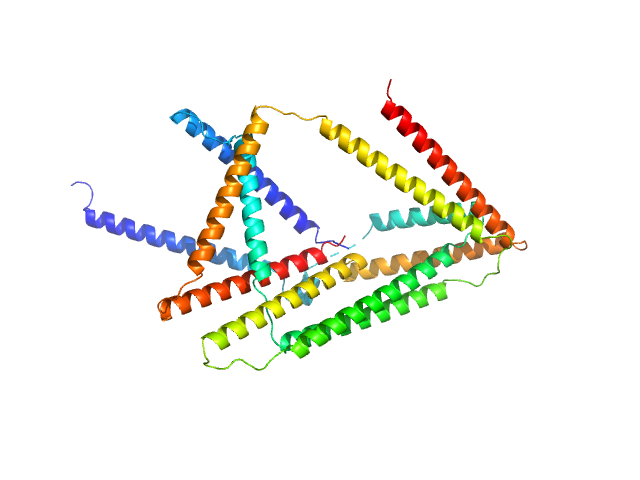
| Sample: |
CyRH1 (cyclic bitrigon from 6 coiled coil segments) monomer, 26 kDa synthetic construct protein
CyRH2 (cyclic bitrigon from 6 coiled coil segments) monomer, 26 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 30
|
Preorganized cyclic modules facilitate the self-assembly of protein nanostructures
Chemical Science (2024)
Snoj J, Lapenta F, Jerala R
|
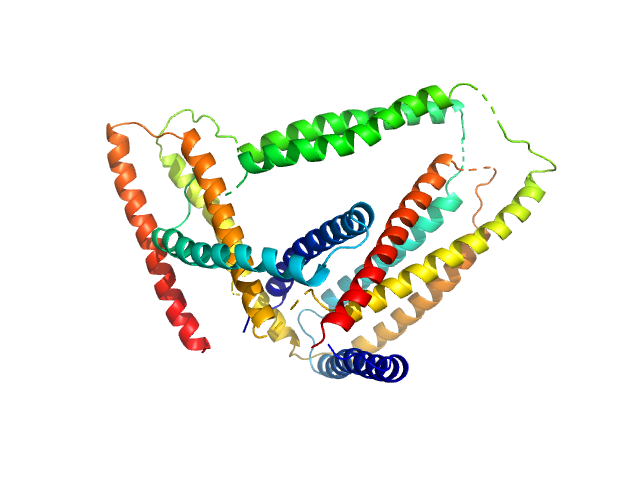
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RH1 (Linear coiled-coil protein origami structure consisting of 6 coiled-coil forming segments) monomer, 26 kDa synthetic construct protein
CyRH2 (cyclic bitrigon from 6 coiled coil segments) monomer, 26 kDa synthetic construct protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Aug 29
|
Preorganized cyclic modules facilitate the self-assembly of protein nanostructures
Chemical Science (2024)
Snoj J, Lapenta F, Jerala R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|
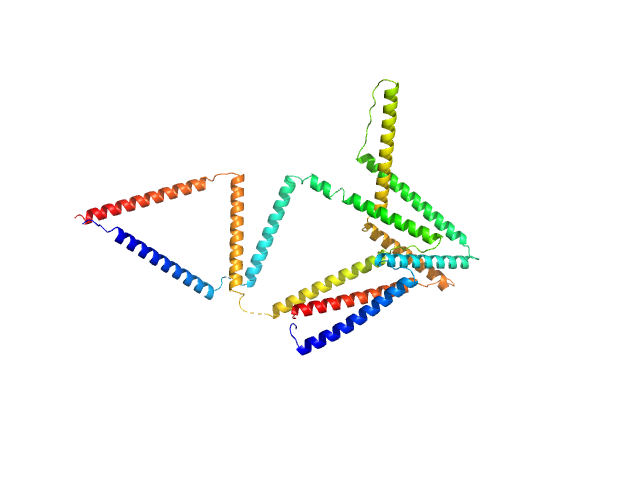
|
| Sample: |
RH1 (Linear coiled-coil protein origami structure consisting of 6 coiled-coil forming segments) monomer, 26 kDa synthetic construct protein
RH2 (Linear coiled-coil protein origami structure consisting of 6 coiled-coil forming segments) monomer, 26 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 9
|
Preorganized cyclic modules facilitate the self-assembly of protein nanostructures
Chemical Science (2024)
Snoj J, Lapenta F, Jerala R
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
161 |
nm3 |
|
|