|
|
|
|
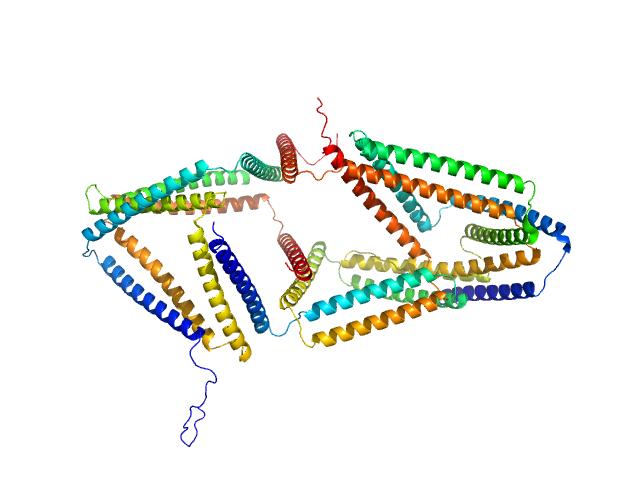
|
| Sample: |
SB6 (Linear coiled-coil protein origami structure consisting of 6 coiled-coil forming segments) monomer, 26 kDa synthetic construct protein
SB9b (Linear coiled-coil protein origami structure consisting of 9 coiled-coil forming segments) monomer, 42 kDa synthetic construct protein
SB9c (Linear coiled-coil protein origami structure consisting of 9 coiled-coil forming segments) monomer, 41 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 25
|
Preorganized cyclic modules facilitate the self-assembly of protein nanostructures
Chemical Science (2024)
Snoj J, Lapenta F, Jerala R
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
443 |
nm3 |
|
|
|
|
|
|
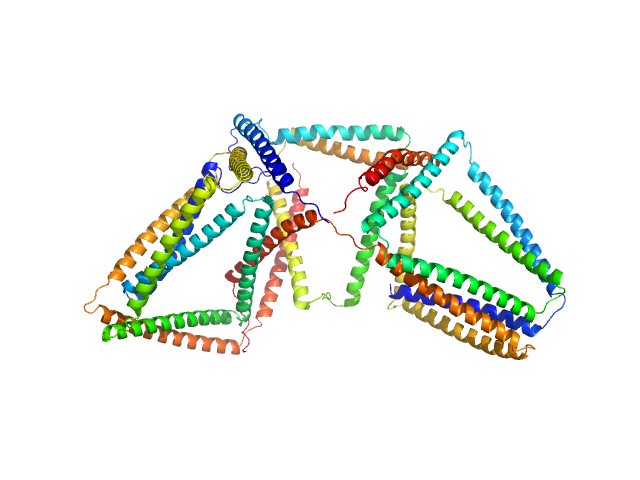
|
| Sample: |
SB9b (Linear coiled-coil protein origami structure consisting of 9 coiled-coil forming segments) monomer, 42 kDa synthetic construct protein
SB9c (Linear coiled-coil protein origami structure consisting of 9 coiled-coil forming segments) monomer, 41 kDa synthetic construct protein
CySB6 (Cyclic coiled-coil protein origami structure consisting of 6 coiled-coil forming segments) monomer, 26 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris, 150 mM NaCL, 3% glycerol, 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 9
|
Preorganized cyclic modules facilitate the self-assembly of protein nanostructures
Chemical Science (2024)
Snoj J, Lapenta F, Jerala R
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
428 |
nm3 |
|
|
|
|
|
|
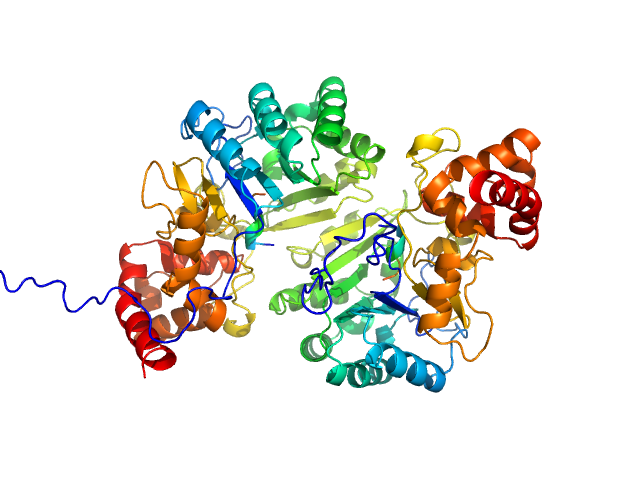
|
| Sample: |
Putative peptide biosynthesis protein YydG dimer, 80 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Nov 30
|
Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat Chem Biol (2023)
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
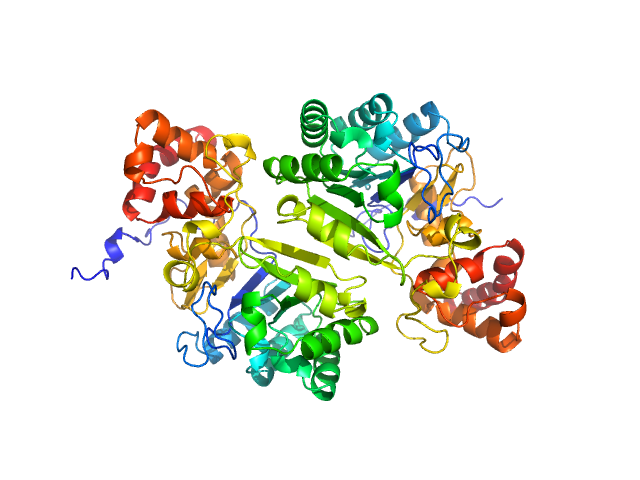
|
| Sample: |
Putative peptide biosynthesis protein YydG dimer, 80 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Nov 29
|
Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat Chem Biol (2023)
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
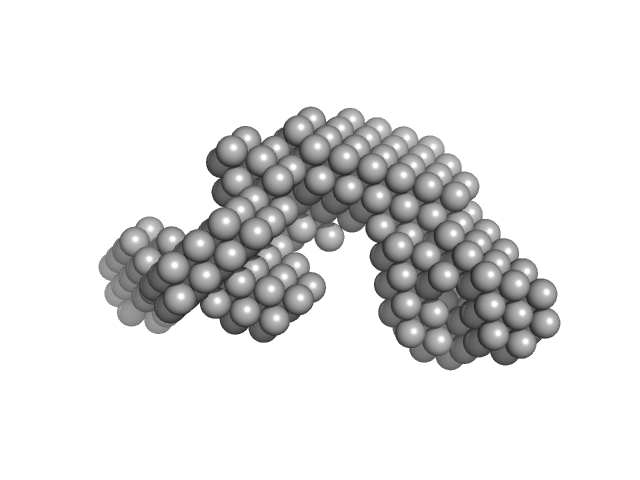
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
296 |
nm3 |
|
|
|
|
|
|
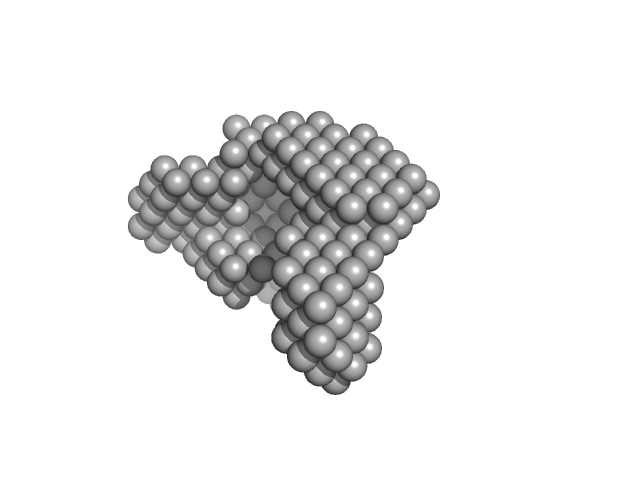
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 336-418 monomer, 9 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
279 |
nm3 |
|
|
|
|
|
|
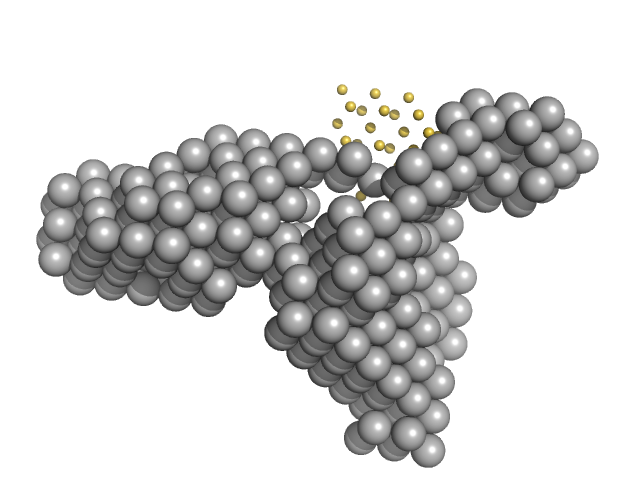
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 225-418 monomer, 21 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
|
|
|
|
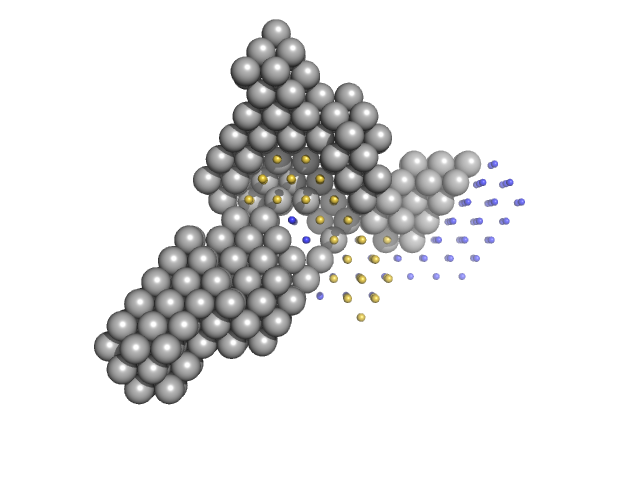
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 225-418 monomer, 21 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 776-898 monomer, 14 kDa Chaetomium thermophilum protein
SMC hinge domain-containing protein, 263-466 monomer, 22 kDa Chaetomium thermophilum protein
SMC hinge domain-containing protein, 1367-1542 monomer, 20 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
355 |
nm3 |
|
|
|
|
|
|
|
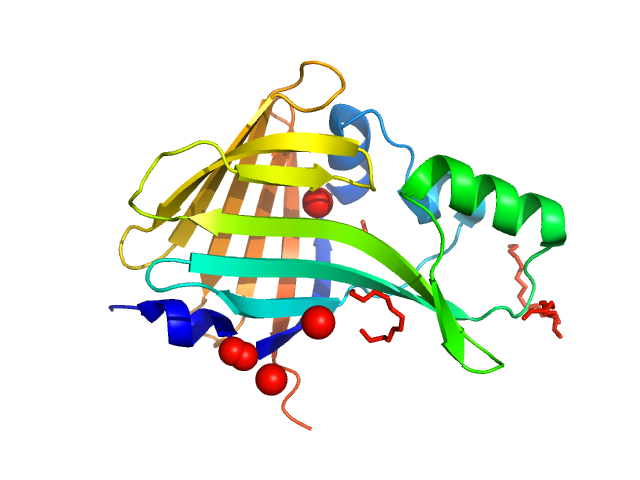
| Sample: |
Oplophorus-luciferin 2-monooxygenase catalytic subunit monomer, 20 kDa Oplophorus gracilirostris protein
|
| Buffer: |
10 mM Tris-HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2021 May 27
|
Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun 14(1):7864 (2023)
Nemergut M, Pluskal D, Horackova J, Sustrova T, Tulis J, Barta T, Baatallah R, Gagnot G, Novakova V, Majerova M, Sedlackova K, Marques SM, Toul M, Damborsky J, Prokop Z, Bednar D, Janin YL, Marek M
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
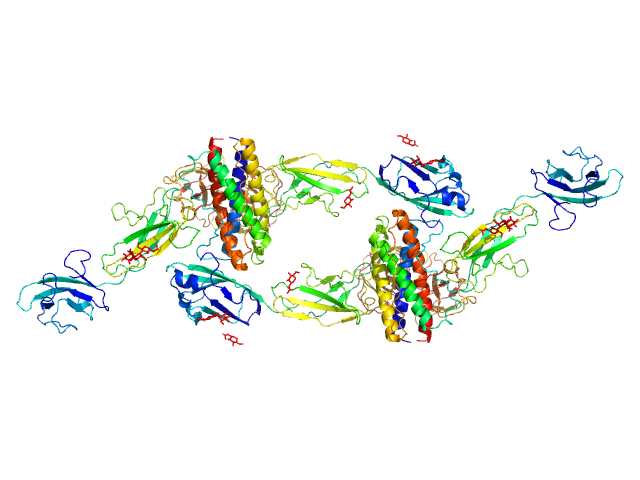
| Sample: |
Interleukin-6 receptor subunit beta dimer, 69 kDa Homo sapiens protein
Interleukin-11 dimer, 36 kDa Homo sapiens protein
Interleukin-11 receptor subunit alpha dimer, 64 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant
Nature Communications 14(1) (2023)
Metcalfe R, Hanssen E, Fung K, Aizel K, Kosasih C, Zlatic C, Doughty L, Morton C, Leis A, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
5.3 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
405 |
nm3 |
|
|