|
|
|
|
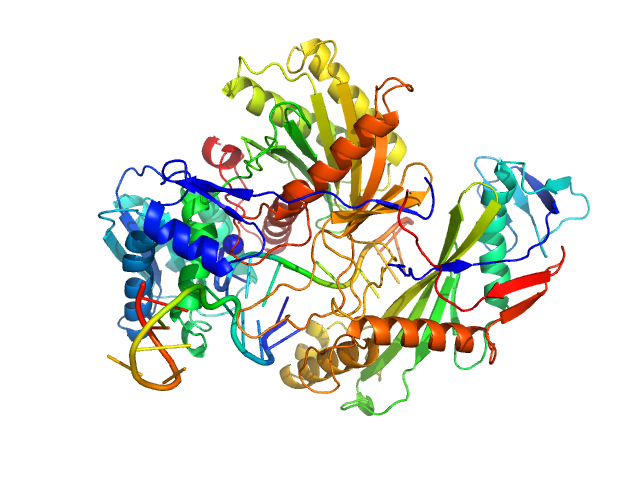
|
| Sample: |
Single-chain full Archaeoglobus fulgidus Argonaute monomer, 78 kDa Archaeoglobus fulgidus protein
5'-end phosphorylated DNA guide strand, 11 nt (MZ864) monomer, 3 kDa DNA
DNA target strand, 11 nt (MZ865) monomer, 3 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Aug 31
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
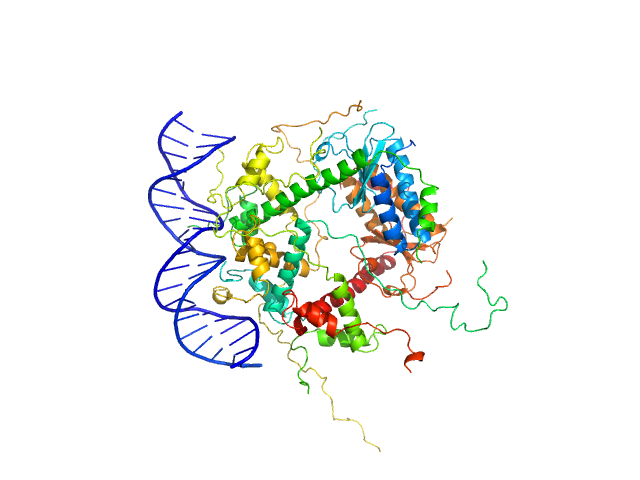
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
21-bp DNA operator fragment monomer, 13 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
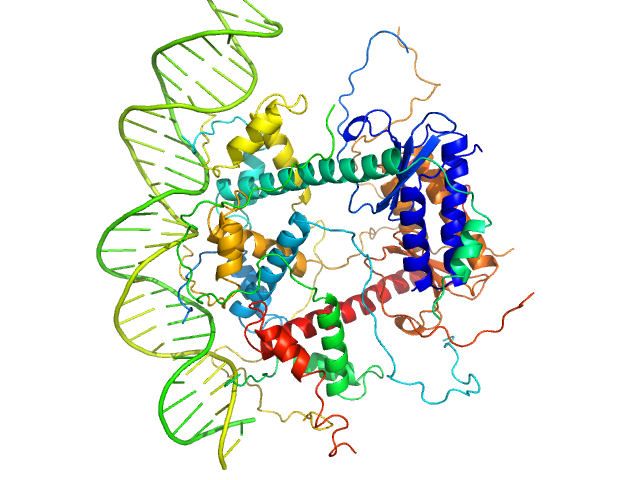
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
31-bp DNA operator box monomer, 19 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
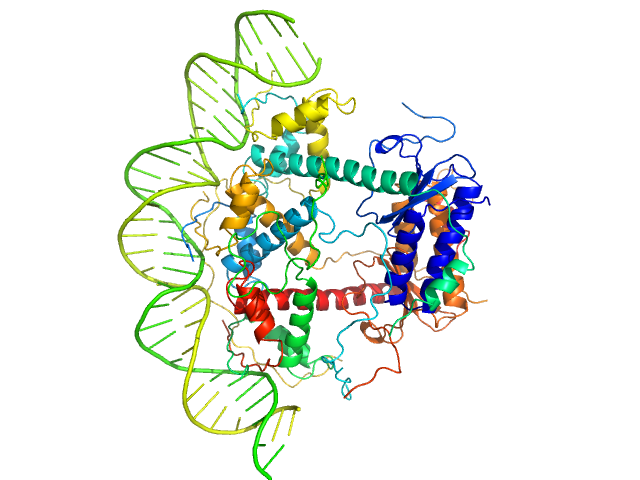
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
33-bp DNA operator fragment monomer, 20 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
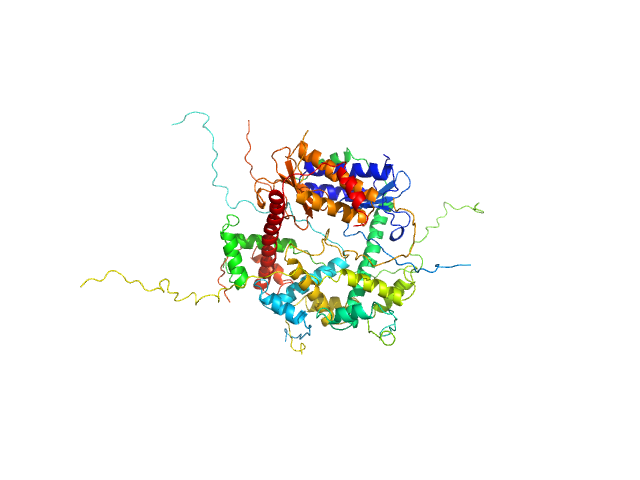
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 6
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
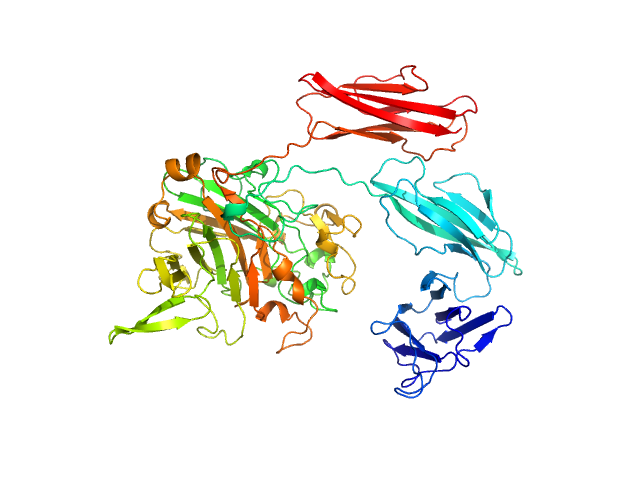
|
| Sample: |
Dockerin type I repeat monomer, 69 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
The Ruminococcus bromii amylosome protein Sas6 binds single and double helical α-glucan structures in starch.
Nat Struct Mol Biol (2024)
Photenhauer AL, Villafuerte-Vega RC, Cerqueira FM, Armbruster KM, Mareček F, Chen T, Wawrzak Z, Hopkins JB, Vander Kooi CW, Janeček Š, Ruotolo BT, Koropatkin NM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
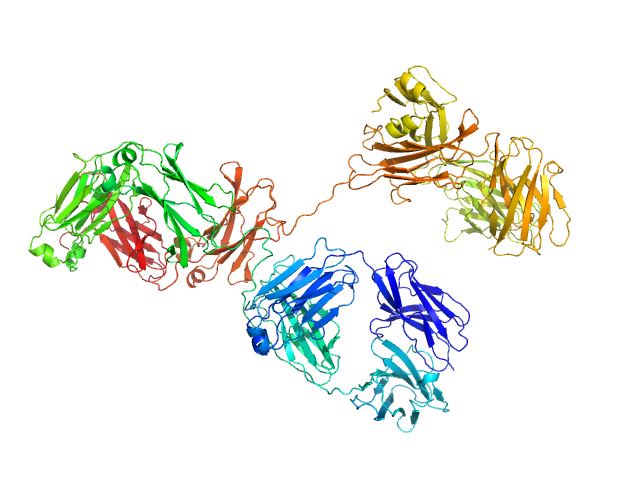
|
| Sample: |
Immunoglobulin G subclass 4 monomer, 145 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 20
|
Using atomistic solution scattering modelling to elucidate the role of the Fc glycans in human IgG4.
PLoS One 19(4):e0300964 (2024)
Spiteri VA, Doutch J, Rambo RP, Bhatt JS, Gor J, Dalby PA, Perkins SJ
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
258 |
nm3 |
|
|
|
|
|
|
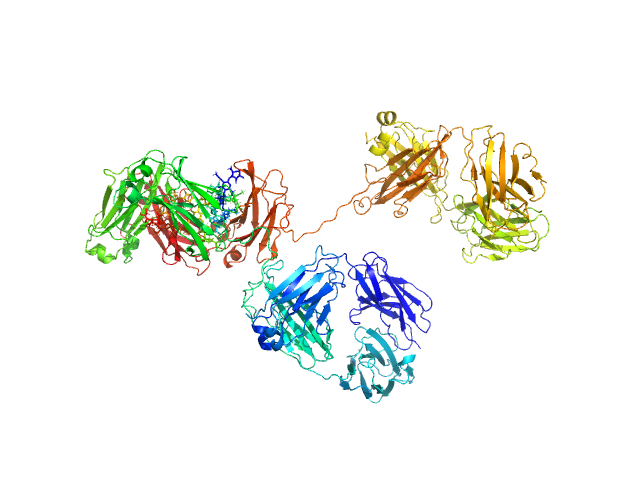
|
| Sample: |
Immunoglobulin G subclass 4 monomer, 145 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 20
|
Using atomistic solution scattering modelling to elucidate the role of the Fc glycans in human IgG4.
PLoS One 19(4):e0300964 (2024)
Spiteri VA, Doutch J, Rambo RP, Bhatt JS, Gor J, Dalby PA, Perkins SJ
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
256 |
nm3 |
|
|
|
|
|
|
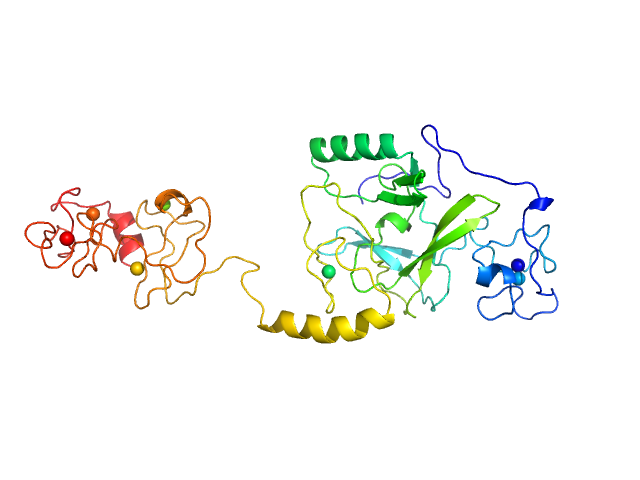
|
| Sample: |
Histone-lysine N-methyltransferase NSD3 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
0.5 M NaCl, 20 mM Tris-HCl, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Nov 25
|
Structural insights into the C-terminus of the histone-lysine N-methyltransferase NSD3 by small-angle X-ray scattering.
Front Mol Biosci 11:1191246 (2024)
Belviso BD, Shen Y, Carrozzini B, Morishita M, di Luccio E, Caliandro R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
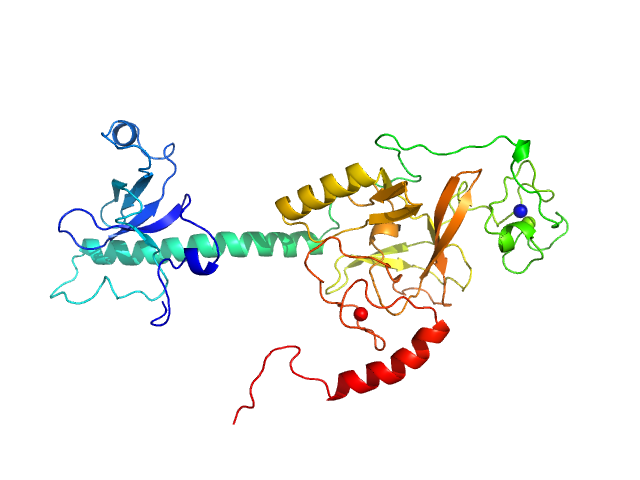
|
| Sample: |
Histone-lysine N-methyltransferase NSD3 monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
0.5 M NaCl, 20 mM Tris-HCl, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Nov 25
|
Structural insights into the C-terminus of the histone-lysine N-methyltransferase NSD3 by small-angle X-ray scattering.
Front Mol Biosci 11:1191246 (2024)
Belviso BD, Shen Y, Carrozzini B, Morishita M, di Luccio E, Caliandro R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
64 |
nm3 |
|
|