|
|
|
|
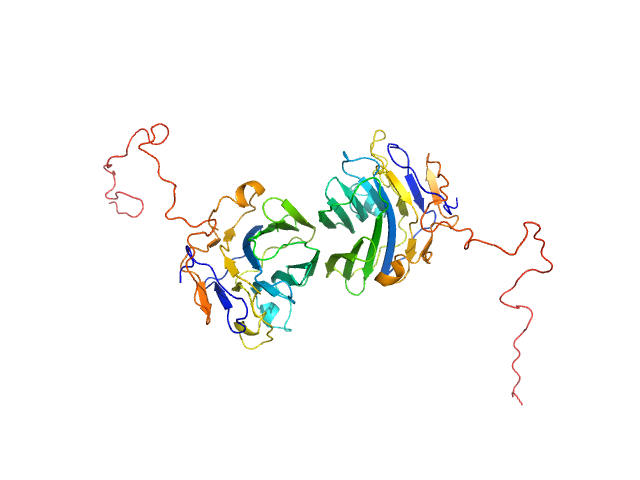
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
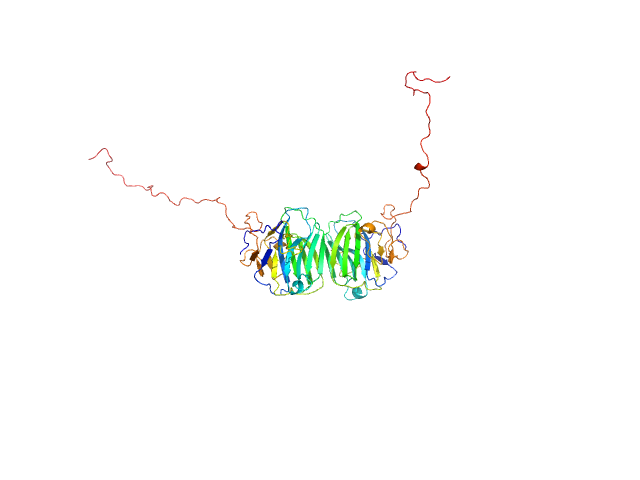
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
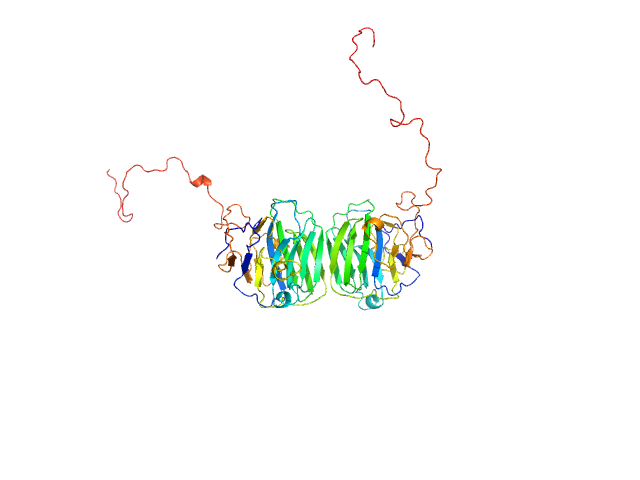
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
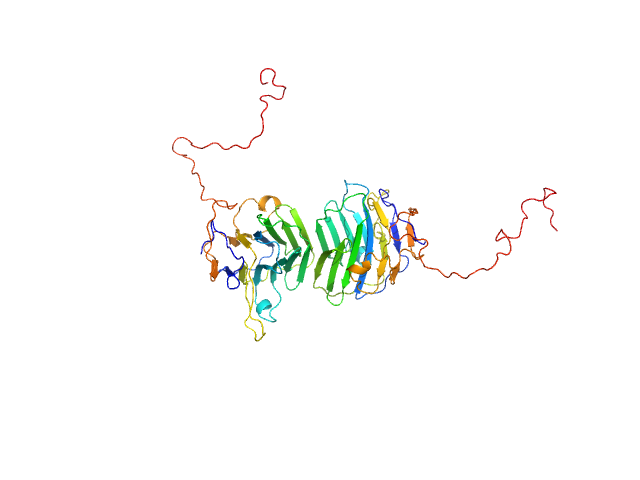
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
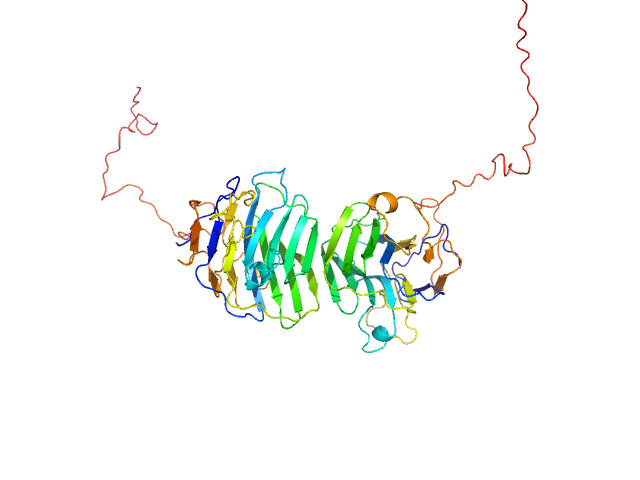
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
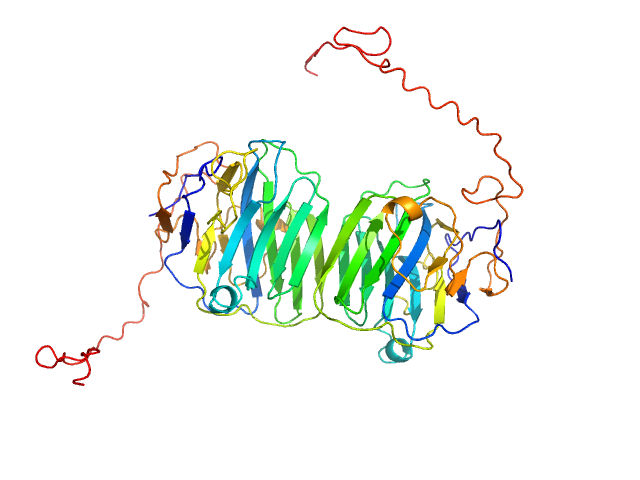
|
| Sample: |
Adhesion G-protein coupled receptor G4 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% (w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 21
|
The dimerized pentraxin-like domain of the adhesion G protein-coupled receptor 112 (ADGRG4) suggests function in sensing mechanical forces.
J Biol Chem :105356 (2023)
Kieslich B, Weiße RH, Brendler J, Ricken A, Schöneberg T, Sträter N
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
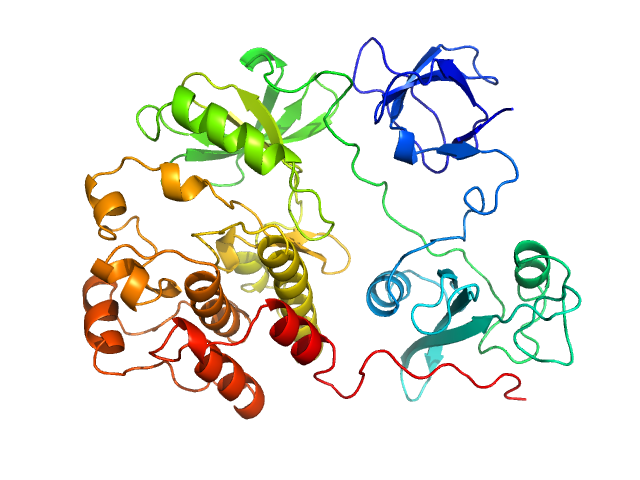
|
| Sample: |
Proto-oncogene tyrosine-protein kinase Src monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 1% glycerol and 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 8
|
An allosteric switch between the activation loop and a c-terminal palindromic phospho-motif controls c-Src function
Nature Communications 14(1) (2023)
Cuesta-Hernández H, Contreras J, Soriano-Maldonado P, Sánchez-Wandelmer J, Yeung W, Martín-Hurtado A, Muñoz I, Kannan N, Llimargas M, Muñoz J, Plaza-Menacho I
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
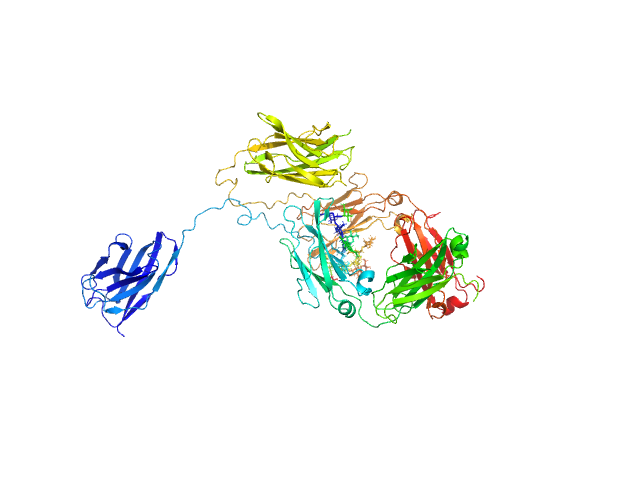
|
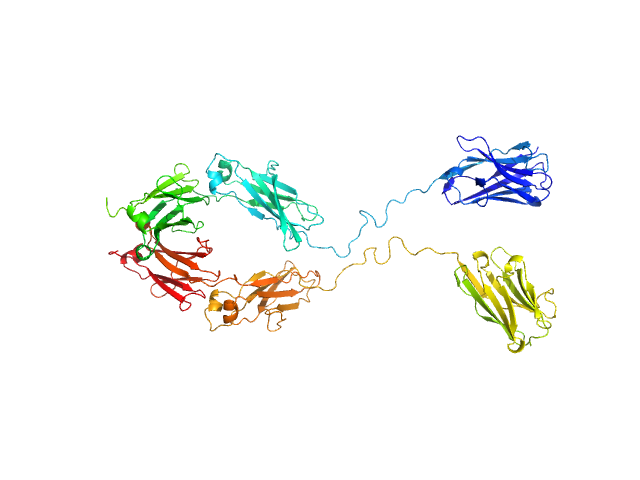
| Sample: |
Immunoglobulin heavy constant gamma 1 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 28
|
The solution structure of the heavy chain-only C5-Fc nanobody reveals exposed variable regions that are optimal for COVID-19 antigen interactions.
J Biol Chem :105337 (2023)
Gao X, Thrush JW, Gor J, Naismith JH, Owens RJ, Perkins SJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
|
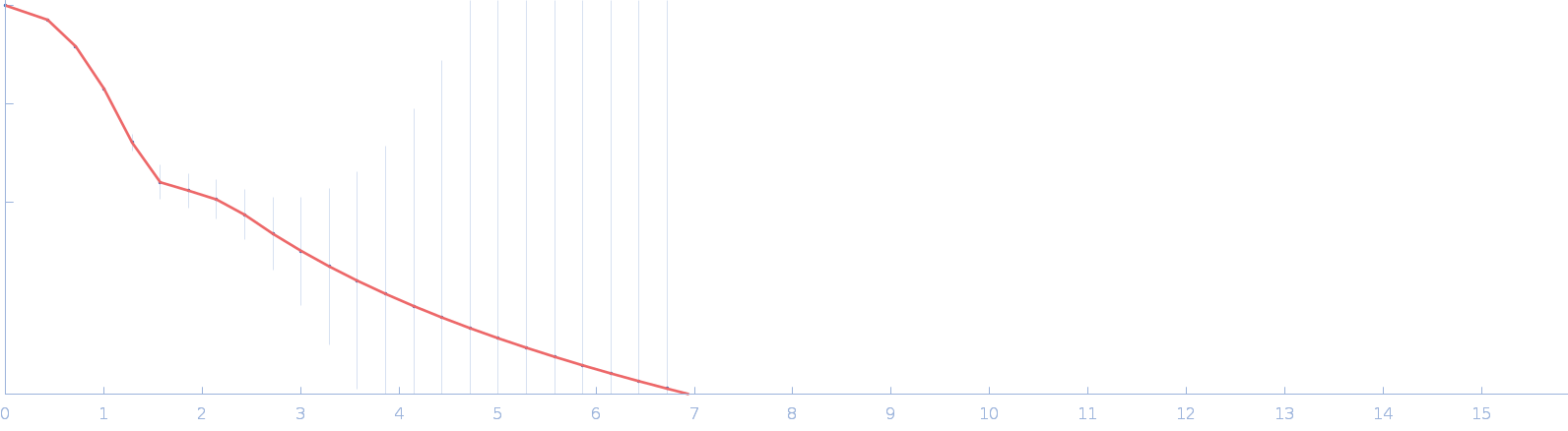
| Sample: |
Immunoglobulin heavy constant gamma 1 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 28
|
The solution structure of the heavy chain-only C5-Fc nanobody reveals exposed variable regions that are optimal for COVID-19 antigen interactions.
J Biol Chem :105337 (2023)
Gao X, Thrush JW, Gor J, Naismith JH, Owens RJ, Perkins SJ
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DUF4374 domain-containing protein monomer, 46 kDa Bacteroides thetaiotaomicron (strain … protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 9
|
Iron acquisition by a commensal bacterium modifies host nutritional immunity during Salmonella infection.
Cell Host Microbe 31(10):1639-1654.e10 (2023)
Spiga L, Fansler RT, Perera YR, Shealy NG, Munneke MJ, David HE, Torres TP, Lemoff A, Ran X, Richardson KL, Pudlo N, Martens EC, Folta-Stogniew E, Yang ZJ, Skaar EP, Byndloss MX, Chazin WJ, Zhu W
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
74 |
nm3 |
|
|