|
|
|
|
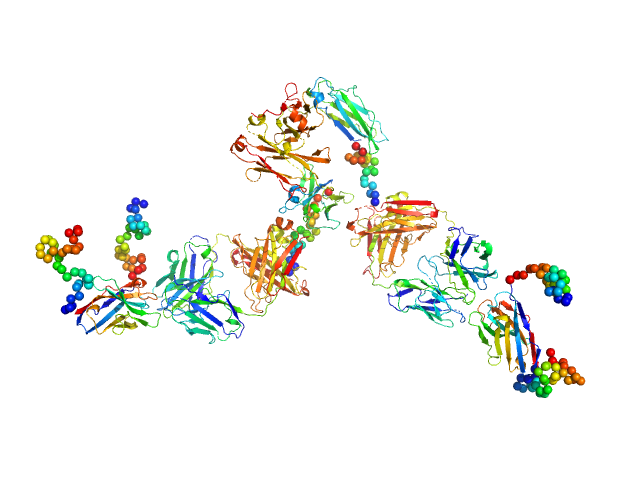
|
| Sample: |
Palivizumab IgG monomer, 145 kDa Commercial (Synagis) protein
Anti-Idiotypic-Palivizumab-Nanobody1 monomer, 20 kDa Lama glama protein
|
| Buffer: |
PBS (137 mM NaCl, 2.7 mM KCl, 12 mM HPO4 2−/H2PO4 −), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Sep 14
|
Respiratory syncytial virus (RSV)-approved monoclonal antibody Palivizumab as ligand for anti-idiotype nanobody-based synthetic cytokine receptors
Journal of Biological Chemistry :105270 (2023)
Ettich J, Wittich C, Moll J, Behnke K, Floss D, Reiners J, Christmann A, Lang P, Smits S, Kolmar H, Scheller J
|
| RgGuinier |
6.0 |
nm |
| Dmax |
20.4 |
nm |
|
|
|
|
|
|
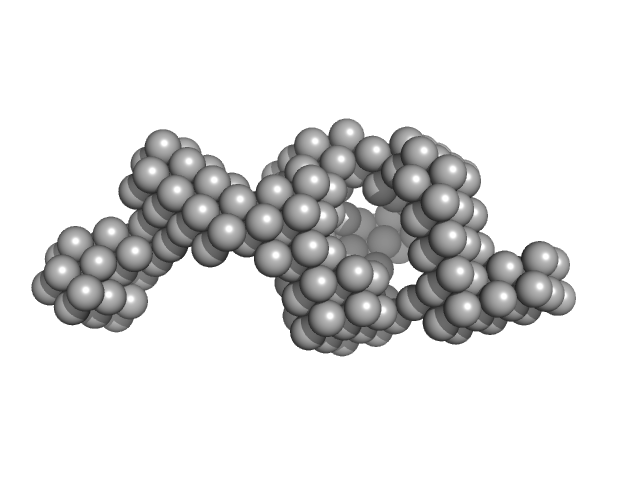
|
| Sample: |
Probable transcriptional regulatory protein (Probably AsnC-family) dimer, 36 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Mar 1
|
Mycobacterium tuberculosis Rv2324 is a multifunctional feast/famine regulatory protein involved in growth, DNA replication and damage control.
Int J Biol Macromol 252:126459 (2023)
Dubey S, Maurya RK, Shree S, Kumar S, Jahan F, Krishnan MY, Ramachandran R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
156 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Probable transcriptional regulatory protein (Probably AsnC-family) octamer, 143 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM EDTA, 2 mM methionine, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Dec 5
|
Mycobacterium tuberculosis Rv2324 is a multifunctional feast/famine regulatory protein involved in growth, DNA replication and damage control.
Int J Biol Macromol 252:126459 (2023)
Dubey S, Maurya RK, Shree S, Kumar S, Jahan F, Krishnan MY, Ramachandran R
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
246 |
nm3 |
|
|
|
|
|
|
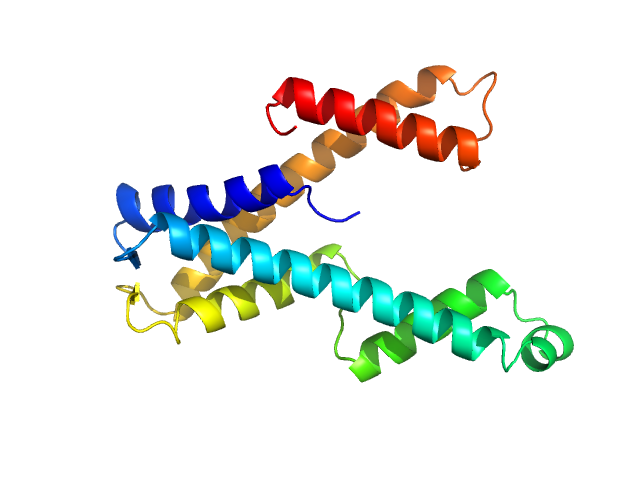
|
| Sample: |
DUF507 family protein monomer, 22 kDa Aquifex aeolicus (strain … protein
|
| Buffer: |
20 mM Hepes pH 7.2, 400 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 7
|
Crystal structure of domain of unknown function 507 (DUF507) reveals a new protein fold.
Sci Rep 13(1):13496 (2023)
McKay CE, Cheng J, Tanner JJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
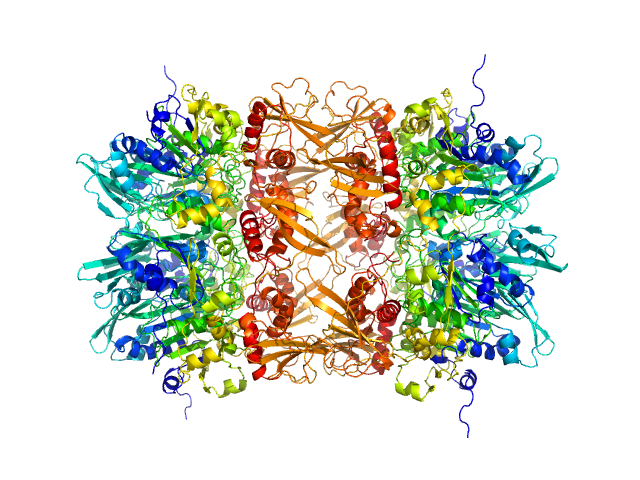
|
| Sample: |
Gp11 encapsidation protein decamer, 446 kDa Lactococcus phage Phi28 protein
|
| Buffer: |
100 mM NaCl, 25 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2015 May 6
|
Lactococcus Phage ASCC Phi28 gp11
Mark White
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.3 |
nm |
|
|
|
|
|
|
|
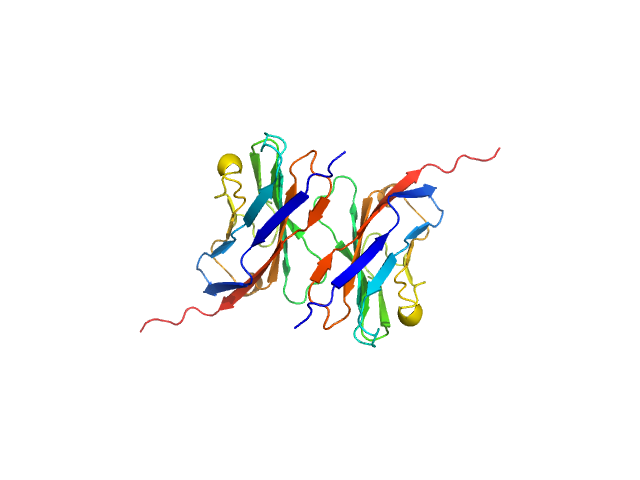
| Sample: |
Myelin protein P0, 15 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaCl, 1 mM EDTA, 20 mM TrisCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jul 21
|
Homomeric interactions of the MPZ Ig domain and their relation to Charcot-Marie-Tooth disease
Brain (2023)
Ptak C, Peterson T, Hopkins J, Ahern C, Shy M, Piper R
|
| RgGuinier |
2.9 |
nm |
| Dmax |
17.0 |
nm |
|
|
|
|
|
|
|
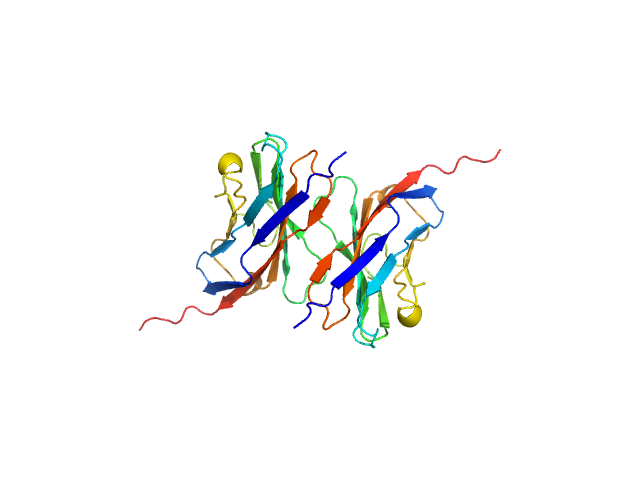
| Sample: |
Myelin protein P0 (W53A, R74A, D75R), 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaCl, 1 mM EDTA, 20 mM TrisCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 14
|
Homomeric interactions of the MPZ Ig domain and their relation to Charcot-Marie-Tooth disease
Brain (2023)
Ptak C, Peterson T, Hopkins J, Ahern C, Shy M, Piper R
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
|
|
|
|
|
|
|
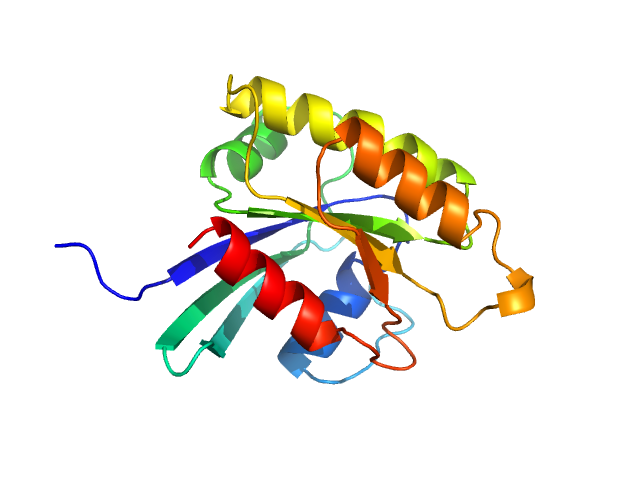
| Sample: |
GTP-binding domain of Ras-like protein 1 monomer, 19 kDa protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% glycerol, 3 mM DTT, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.1 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
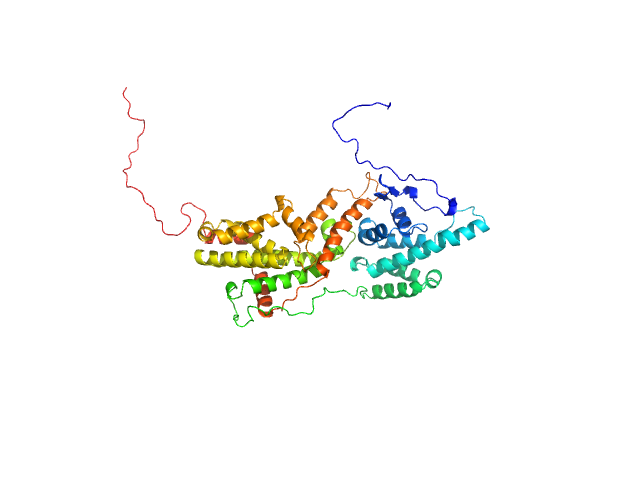
| Sample: |
Cell division control protein 25 monomer, 55 kDa Candida albicans (strain … protein
|
| Buffer: |
20 mM Na-phosphate, 150 mM NaCl, 5% glycerol, 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
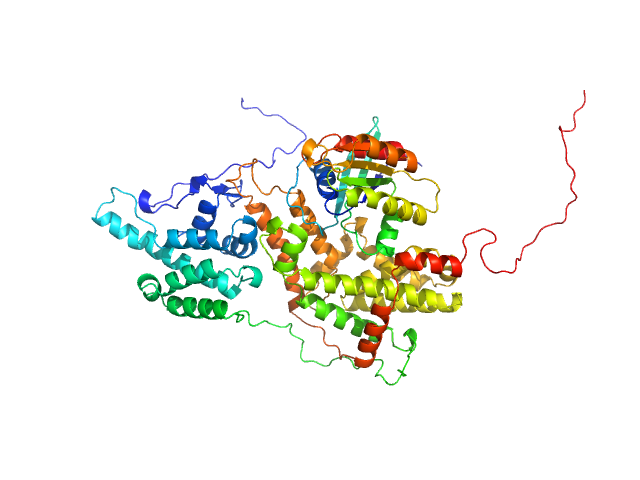
|
| Sample: |
GTP-binding domain of Ras-like protein 1 monomer, 19 kDa protein
Cell division control protein 25 monomer, 55 kDa Candida albicans (strain … protein
|
| Buffer: |
20 mM Na-phosphate, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
99 |
nm3 |
|
|