|
|
|
|

|
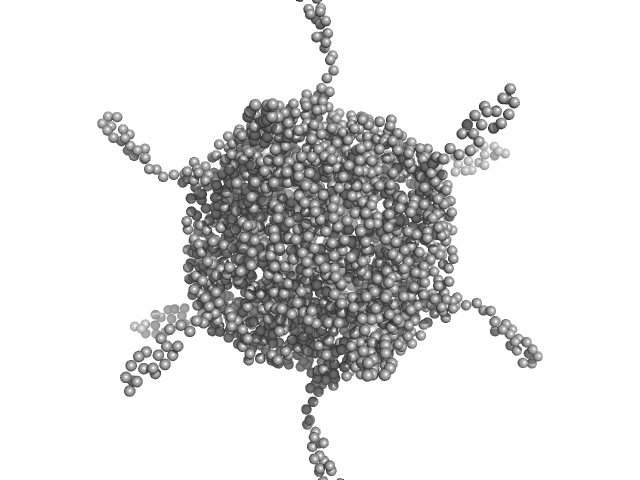
| Sample: |
DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
409 |
nm3 |
|
|
|
|
|
|
|
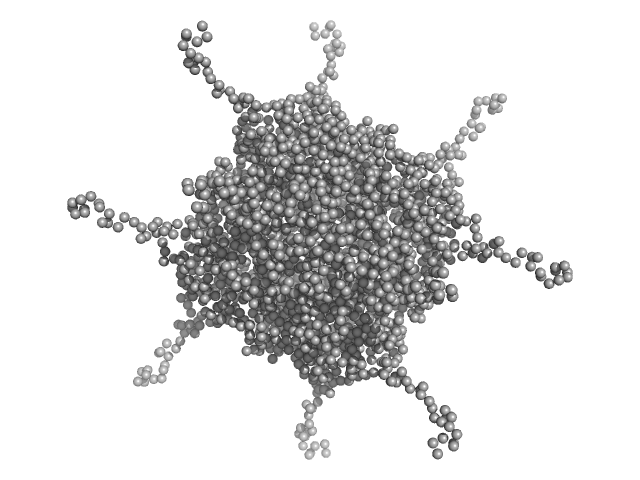
| Sample: |
DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
20.6 |
nm |
| VolumePorod |
404 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation, DPS-D43A (Ferritin superfamily) dodecamer D43A mutation dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
431 |
nm3 |
|
|
|
|
|
|
|
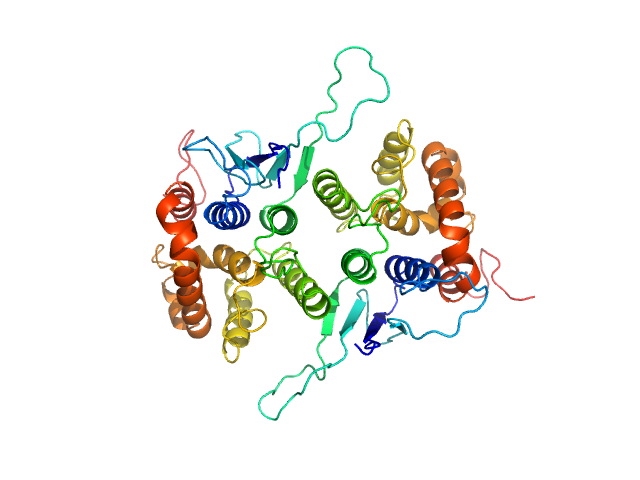
| Sample: |
Glutamate--tRNA ligase dimer, 57 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
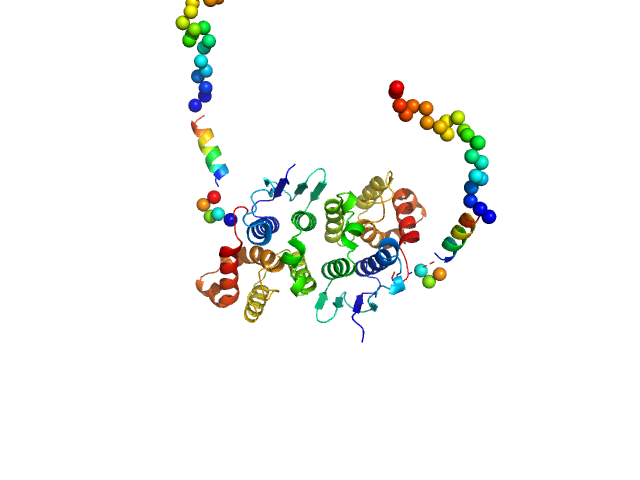
|
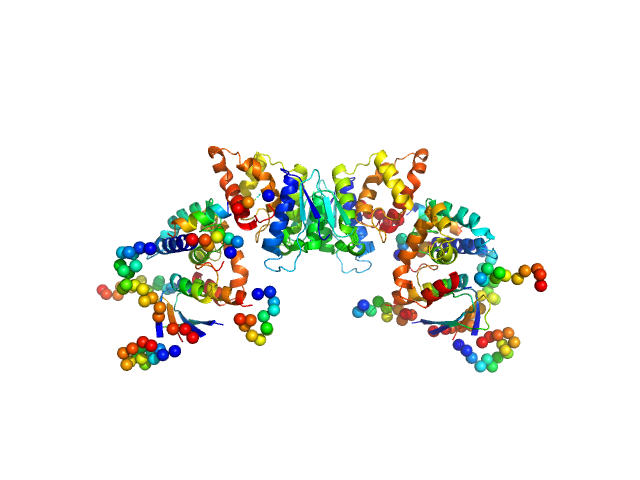
| Sample: |
TRNA import protein tRIP dimer, 49 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Chimeric construct containing the GST-like domains of P. berghei tRNA import protein and glutamyl-tRNA synthetase dimer, 105 kDa Plasmodium berghei ANKA protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
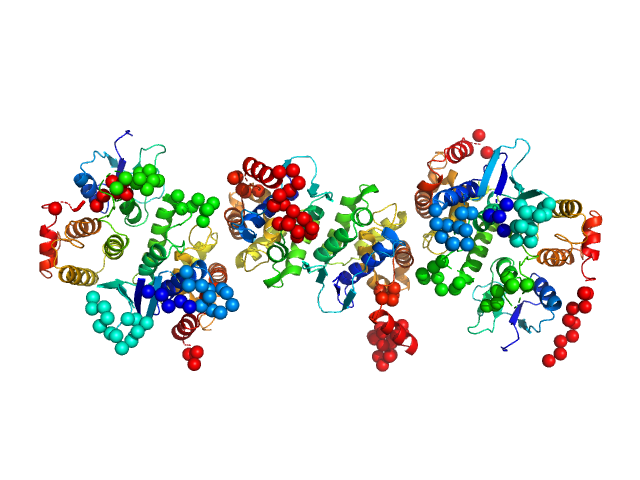
| Sample: |
Glutamine--tRNA ligase dimer, 46 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
TRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|
|
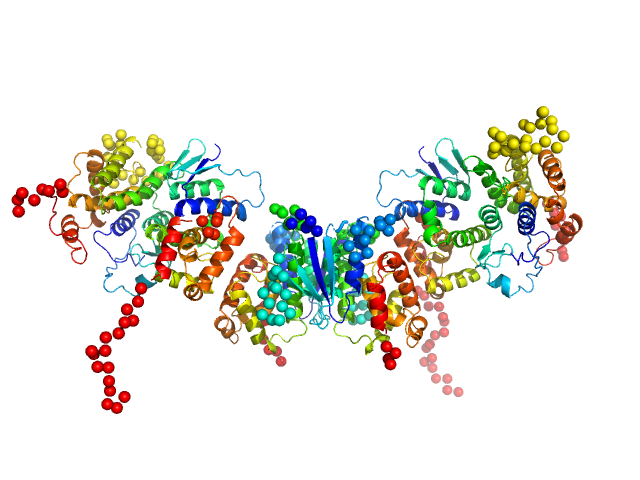
| Sample: |
Methionine--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
TRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|
|
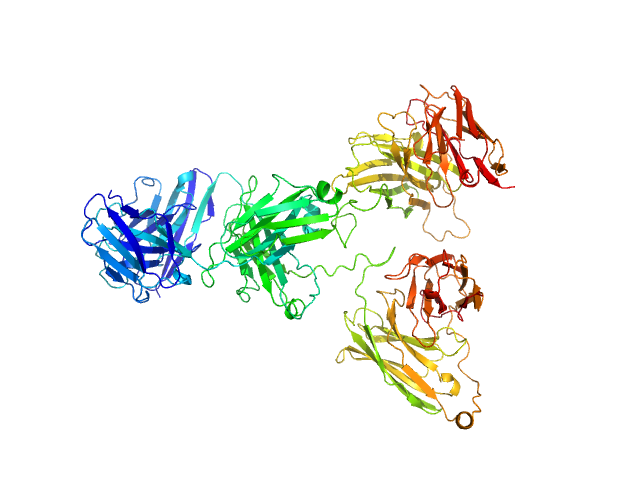
| Sample: |
Tri-specific antibody monomer, 102 kDa Homo sapiens protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 26
|
TRYBE®: an Fc-free antibody format with three monovalent targeting arms engineered for long in vivo half-life.
MAbs 15(1):2160229 (2023)
Davé E, Durrant O, Dhami N, Compson J, Broadbridge J, Archer S, Maroof A, Whale K, Menochet K, Bonnaillie P, Barry E, Wild G, Peerboom C, Bhatta P, Ellis M, Hinchliffe M, Humphreys DP, Heywood SP
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fusion protein of LSm and MyoX-coil, 1066 kDa Artificial protein protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 May 23
|
Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J Am Chem Soc (2022)
Ohara N, Kawakami N, Arai R, Adachi N, Moriya T, Kawasaki M, Miyamoto K
|
| RgGuinier |
9.1 |
nm |
| Dmax |
22.7 |
nm |
|
|