|
|
|
|
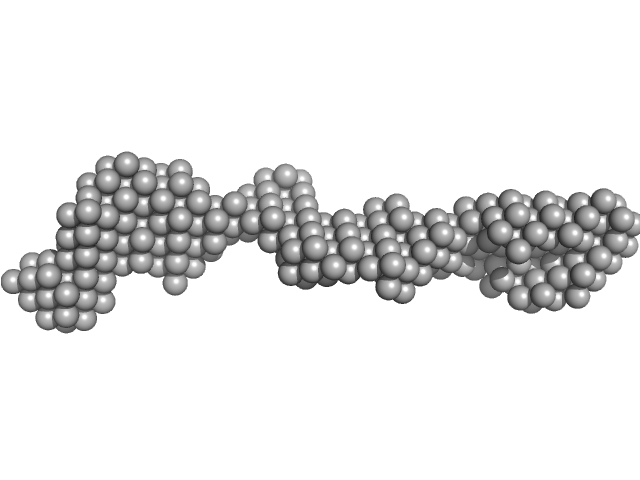
|
| Sample: |
Receptor-type tyrosine-protein phosphatase mu monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES, 250 mM NaCl, 3% v/v glycerol,, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 13
|
Determinants of receptor tyrosine phosphatase homophilic adhesion: structural comparison of PTPRK and PTPRM extracellular domains
Journal of Biological Chemistry :102750 (2022)
Hay I, Shamin M, Caroe E, Mohammed A, Svergun D, Jeffries C, Graham S, Sharpe H, Deane J
|
| RgGuinier |
7.2 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
|
|
|
|
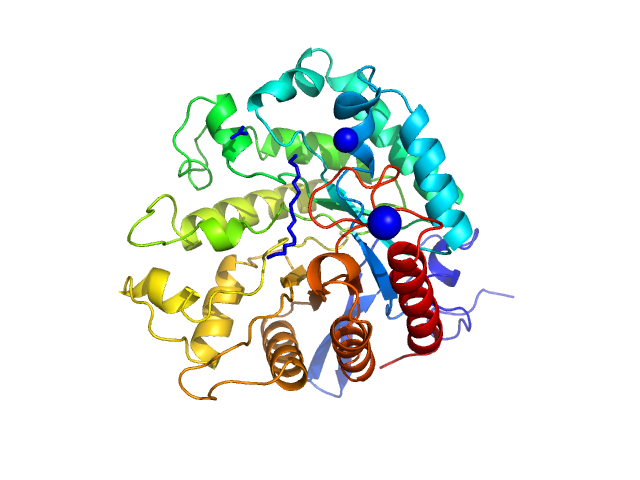
|
| Sample: |
GH5 endo-mannanase from Capybara gut metagenome monomer, 46 kDa metagenome protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 Oct 30
|
Glycoside hydrolase subfamily GH5_57 features a highly redesigned catalytic interface to process complex hetero-β-mannans.
Acta Crystallogr D Struct Biol 78(Pt 11):1358-1372 (2022)
Martins MP, Morais MAB, Persinoti GF, Galinari RH, Yu L, Yoshimi Y, Passos Nunes FB, Lima TB, Barbieri SF, Silveira JLM, Lombard V, Terrapon N, Dupree P, Henrissat B, Murakami MT
|
| RgGuinier |
2.3 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
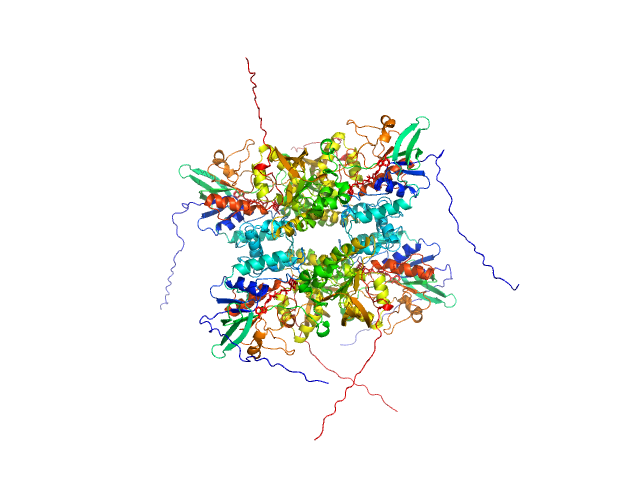
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
394 |
nm3 |
|
|
|
|
|
|
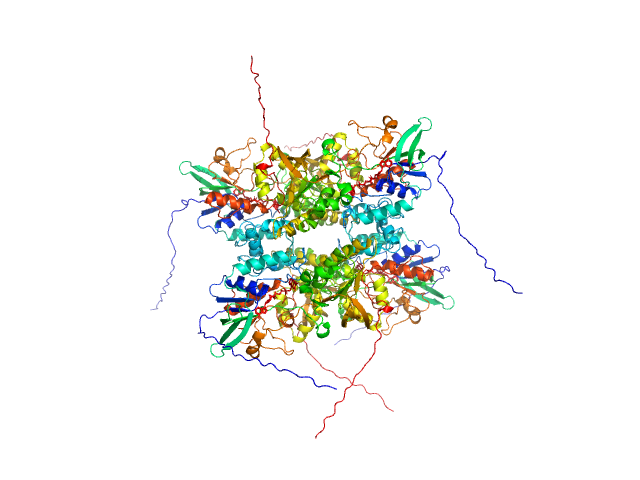
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|
|
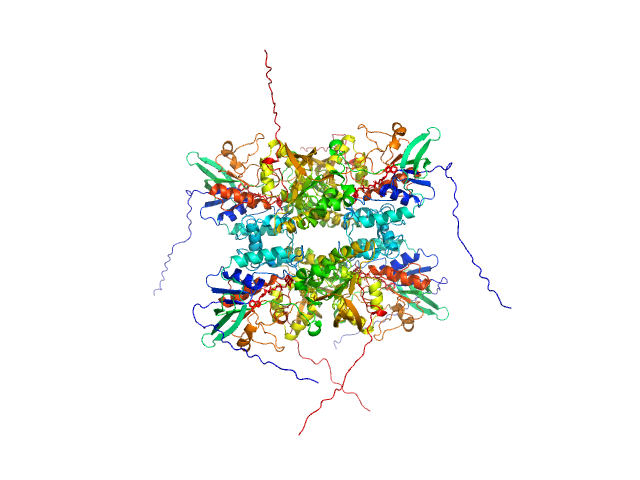
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|
|
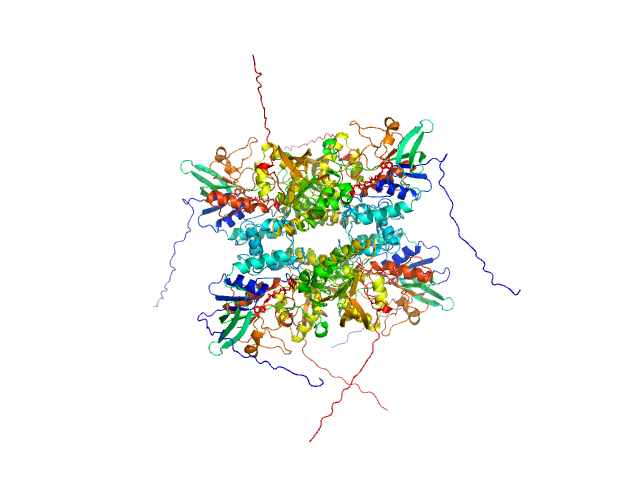
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|
|
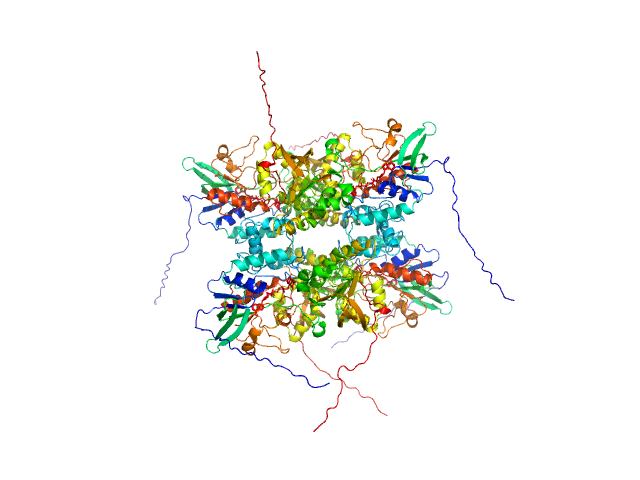
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|
|
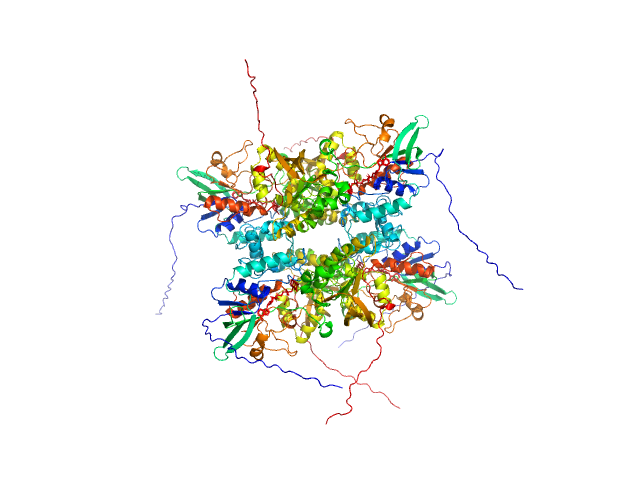
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
396 |
nm3 |
|
|
|
|
|
|
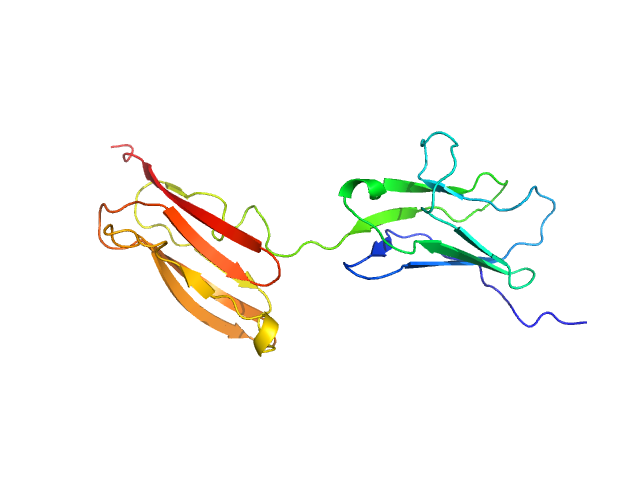
|
| Sample: |
Obscurin Ig domains 12/13 short monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 20
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
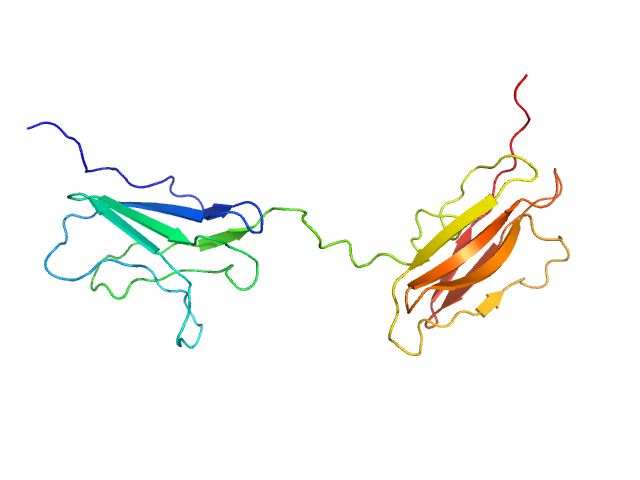
|
| Sample: |
Obscurin Ig domains 12/13 long monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 20
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|