|
|
|
|
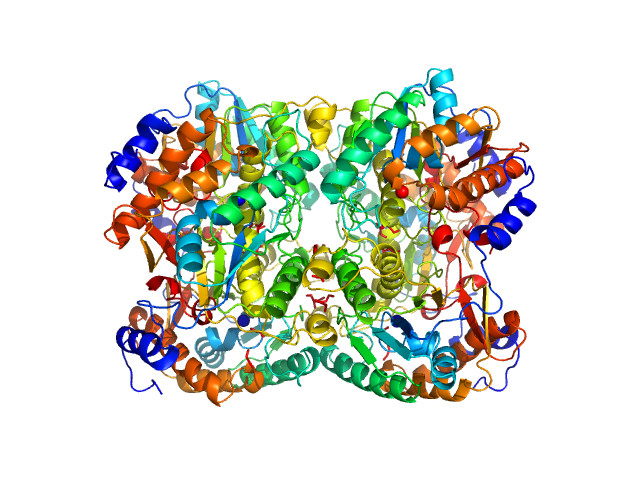
|
| Sample: |
Diacetylchitobiose deacetylase hexamer, 186 kDa Thermococcus chitonophagus protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Sep 26
|
Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci 23(24) (2022)
Biniek-Antosiak K, Bejger M, Śliwiak J, Baranowski D, Mohammed ASA, Svergun DI, Rypniewski W
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
317 |
nm3 |
|
|
|
|
|
|
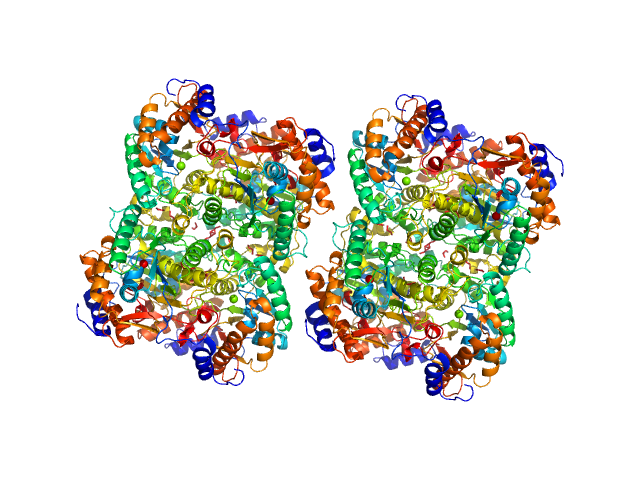
|
| Sample: |
Diacetylchitobiose deacetylase, 185 kDa Thermococcus chitonophagus protein
|
| Buffer: |
20 mM TRIS, 200 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Sep 26
|
Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci 23(24) (2022)
Biniek-Antosiak K, Bejger M, Śliwiak J, Baranowski D, Mohammed ASA, Svergun DI, Rypniewski W
|
|
|
|
|
|
|
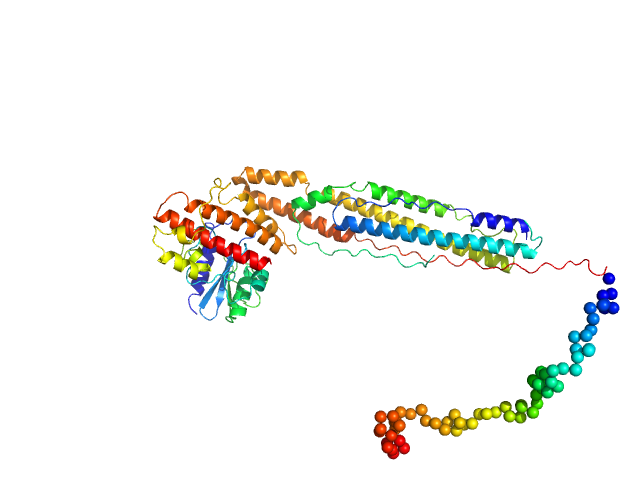
|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 27 kDa Mycobacterium tuberculosis (strain … protein
ESX-1 secretion-associated protein EspB monomer, 37 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 12
|
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
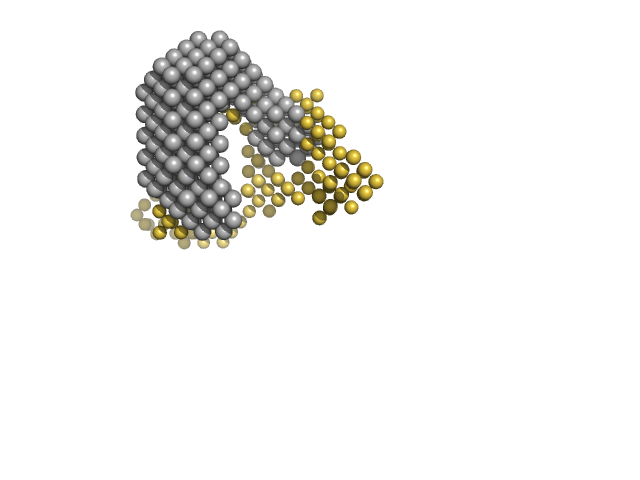
|
| Sample: |
Multidrug resistance operon repressor dimer, 32 kDa Pseudomonas aeruginosa protein
34 base pair double-stranded DNA monomer, 21 kDa synthetic construct DNA
|
| Buffer: |
20mM NaPO4, 150 mM NaCl, 10 mM DTT, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 May 30
|
Small-angle X-ray and neutron scattering of MexR and its complex with DNA supports a conformational selection binding model
Biophysical Journal (2022)
Caporaletti F, Pietras Z, Morad V, Mårtensson L, Gabel F, Wallner B, Martel A, Sunnerhagen M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
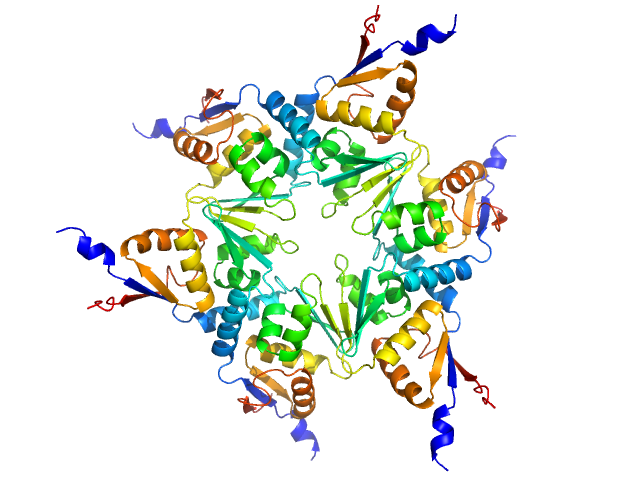
|
| Sample: |
Longitudinals lacking protein, isoform G hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
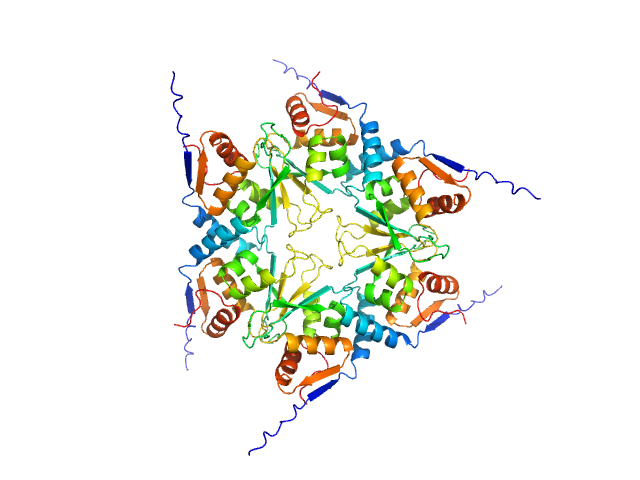
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
167 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
Rho GTPase-activating protein 35 monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8 350 mM NaCl 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 11
|
Tandem engagement of phosphotyrosines by the dual SH2 domains of p120RasGAP.
Structure (2022)
Stiegler AL, Vish KJ, Boggon TJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
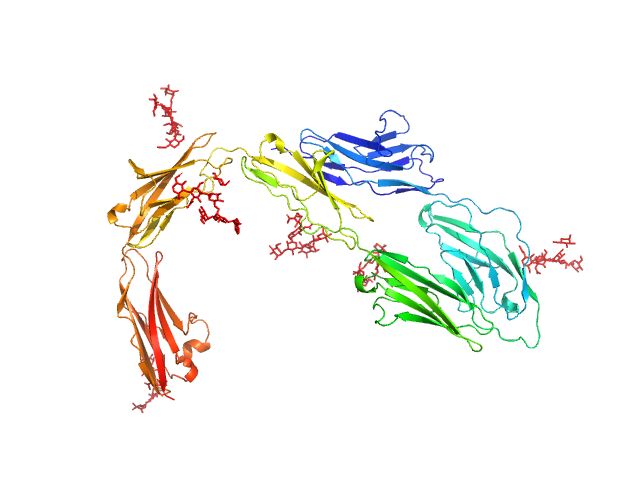
|
| Sample: |
Contactin-1 I433V dimer, 134 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
4.9 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
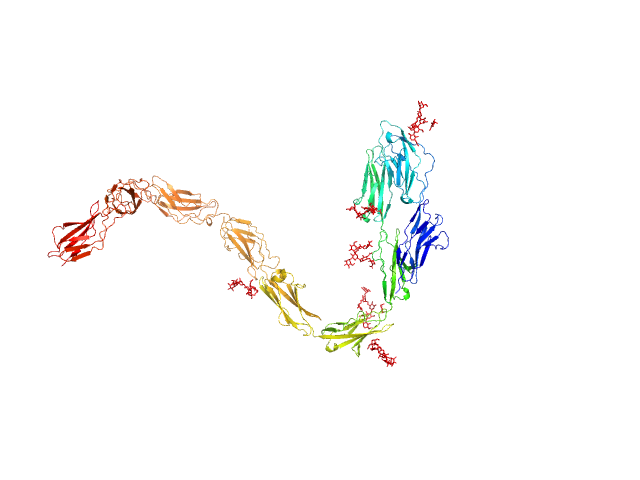
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
|
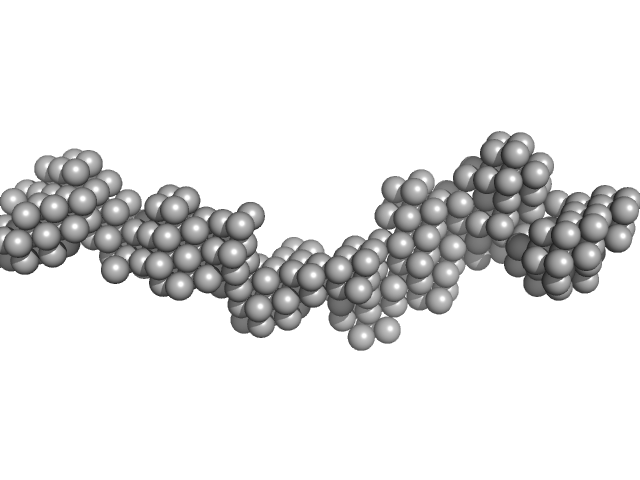
| Sample: |
Receptor-type tyrosine-protein phosphatase kappa monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES, 250 mM NaCl, 3% v/v glycerol,, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 13
|
Determinants of receptor tyrosine phosphatase homophilic adhesion: structural comparison of PTPRK and PTPRM extracellular domains
Journal of Biological Chemistry :102750 (2022)
Hay I, Shamin M, Caroe E, Mohammed A, Svergun D, Jeffries C, Graham S, Sharpe H, Deane J
|
| RgGuinier |
7.0 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
252 |
nm3 |
|
|