|
|
|
|
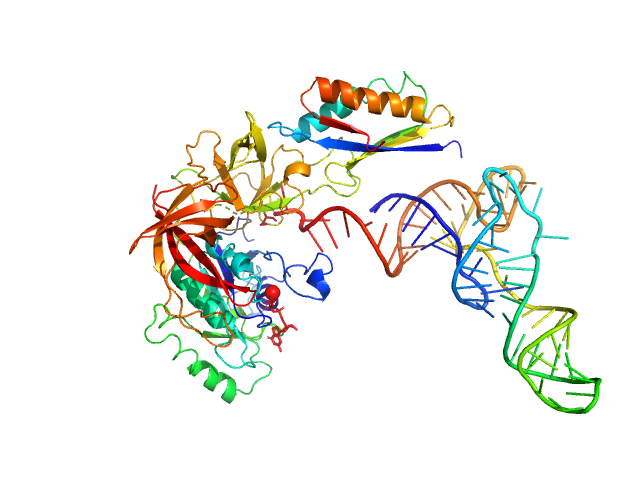
|
| Sample: |
Translation initiation factor 2 subunit gamma monomer, 46 kDa Saccharolobus solfataricus (strain … protein
Translation initiation factor 2 subunit alpha monomer, 10 kDa Saccharolobus solfataricus (strain … protein
Transfer RNA monomer, 23 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS- NaOH pH 6.7, 200 mM NaCl, 5 mM MgCl 2, 1 mM GDPNP, pH: 6.7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
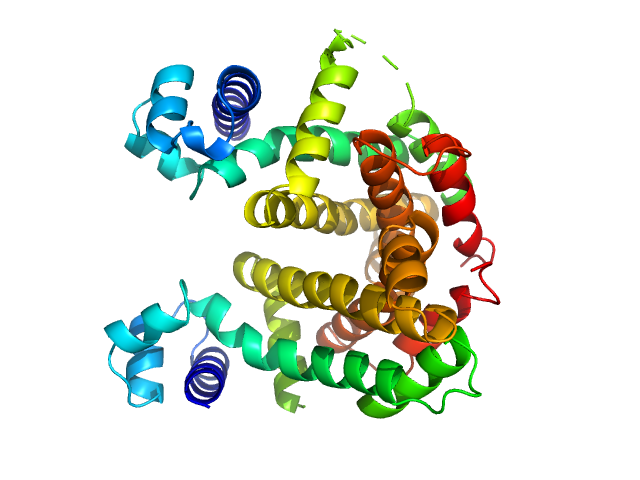
|
| Sample: |
Putative tetR-family transcriptional regulator dimer, 47 kDa Streptomyces coelicolor (strain … protein
|
| Buffer: |
20 mM Tris/HCl, 400 mM NaCl, 50 mM imidazole, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 May 2
|
Crystal Structures of Free and Ligand-Bound Forms of the TetR/AcrR-Like Regulator SCO3201 from Streptomyces coelicolor Suggest a Novel Allosteric Mechanism.
FEBS J (2022)
Werten S, Waack P, Palm GJ, Virolle MJ, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
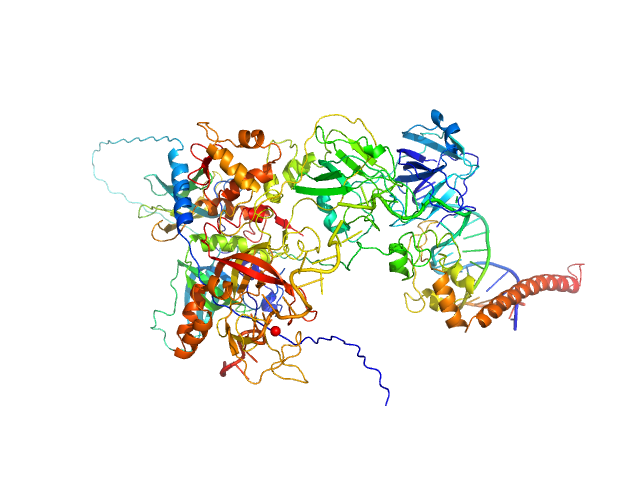
|
| Sample: |
DNA repair protein complementing XP-A cells monomer, 27 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 49 kDa Homo sapiens protein
Replication protein A 32 kDa subunit monomer, 25 kDa Homo sapiens protein
Replication protein A 14 kDa subunit monomer, 14 kDa Homo sapiens protein
3-prime ss-ds DNA junction NER model substrate monomer, 17 kDa DNA
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Mar 4
|
Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair
Proceedings of the National Academy of Sciences 119(34) (2022)
Kim M, Kim H, D’Souza A, Gallagher K, Jeong E, Topolska-Wós A, Ogorodnik Le Meur K, Tsai C, Tsai M, Kee M, Tainer J, Yeo J, Chazin W, Schärer O
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
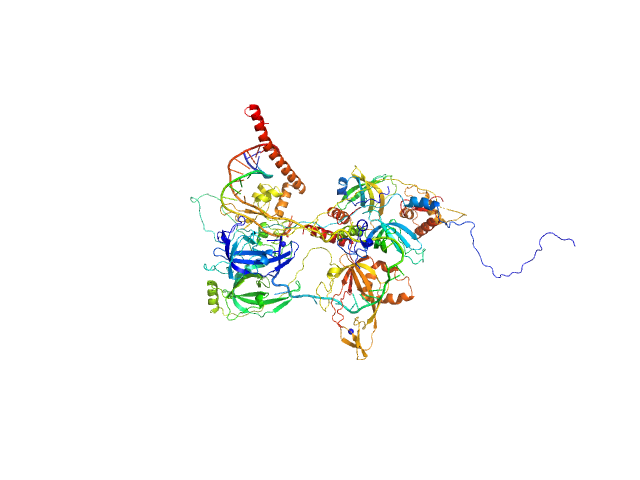
|
| Sample: |
Replication protein A 14 kDa subunit monomer, 14 kDa Homo sapiens protein
DNA repair protein complementing XP-A cells monomer, 27 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 49 kDa Homo sapiens protein
Replication protein A 32 kDa subunit monomer, 25 kDa Homo sapiens protein
5-prime ss-ds DNA junction NER model substrate monomer, 17 kDa DNA
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Mar 4
|
Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair
Proceedings of the National Academy of Sciences 119(34) (2022)
Kim M, Kim H, D’Souza A, Gallagher K, Jeong E, Topolska-Wós A, Ogorodnik Le Meur K, Tsai C, Tsai M, Kee M, Tainer J, Yeo J, Chazin W, Schärer O
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
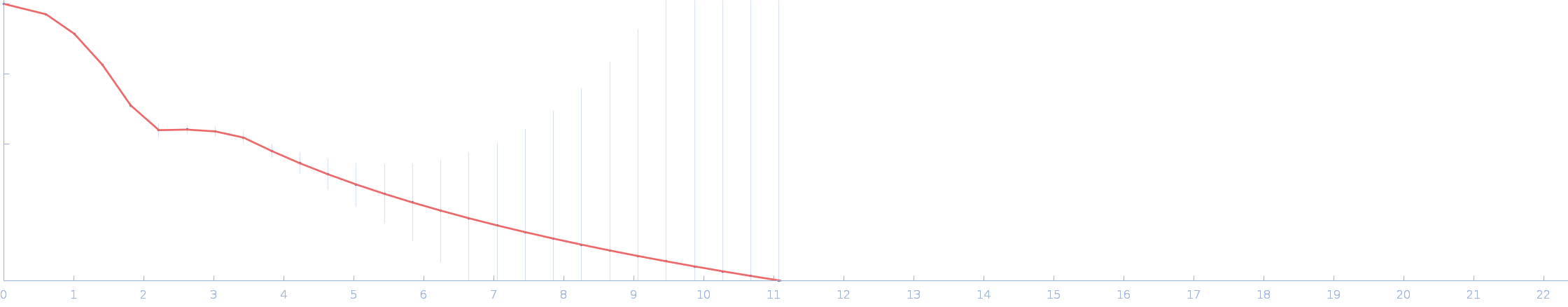
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Calmidazolium monomer, 1 kDa
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rhoptry kinase family protein tetramer, 143 kDa Toxoplasma gondii (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 19
|
Divergent kinase WNG1 is regulated by phosphorylation of an atypical activation sub-domain.
Biochem J (2022)
Dewangan PS, Beraki TG, Paiz EA, Appiah Mensah D, Chen Z, Reese ML
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|
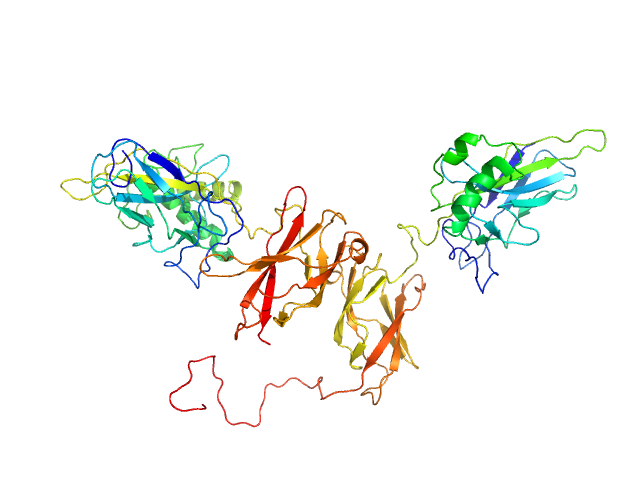
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
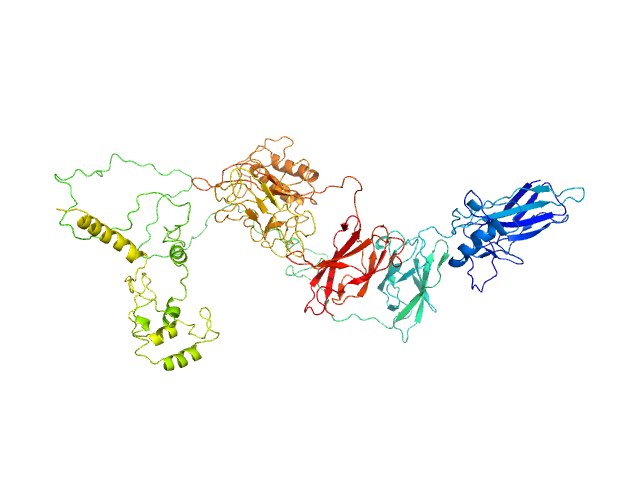
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
|
|
|
|
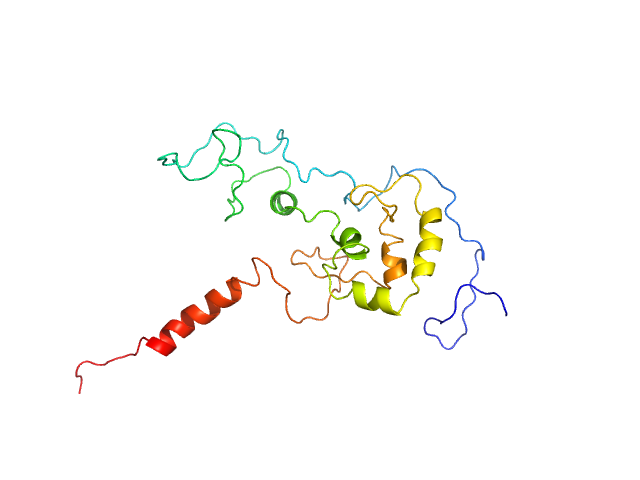
|
| Sample: |
Transcription factor p65 340-549 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein DPCD monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
Deciphering cellular and molecular determinants of human DPCD protein in complex with RUVBL1/RUVBL2 AAA-ATPases.
J Mol Biol :167760 (2022)
Dos Santos Morais R, Santo PE, Ley M, Schelcher C, Abel Y, Plassart L, Deslignière E, Chagot ME, Quinternet M, Paiva ACF, Hessmann S, Morellet N, M F Sousa P, Vandermoere F, Bertrand E, Charpentier B, Bandeiras TM, Plisson-Chastang C, Verheggen C, Cianférani S, Manival X
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
39 |
nm3 |
|
|