|
|
|
|
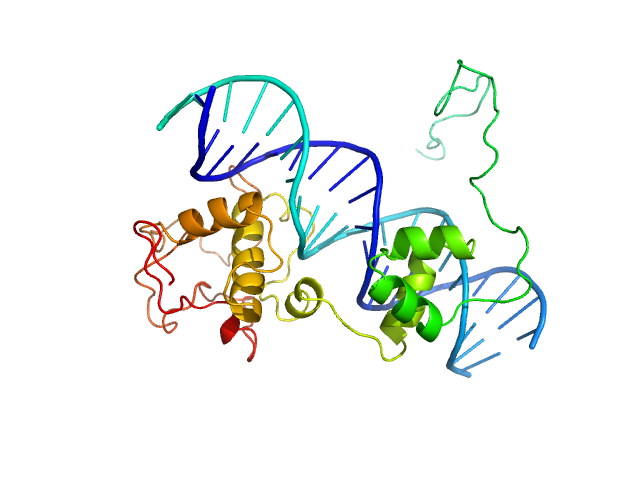
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
21mer dsDNA monomer, 13 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
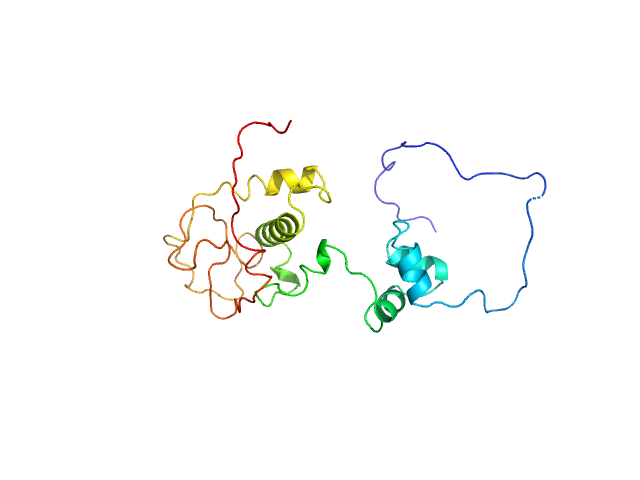
|
| Sample: |
Immunity repressor monomer, 24 kDa Mycobacterium phage TipsytheTRex protein
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
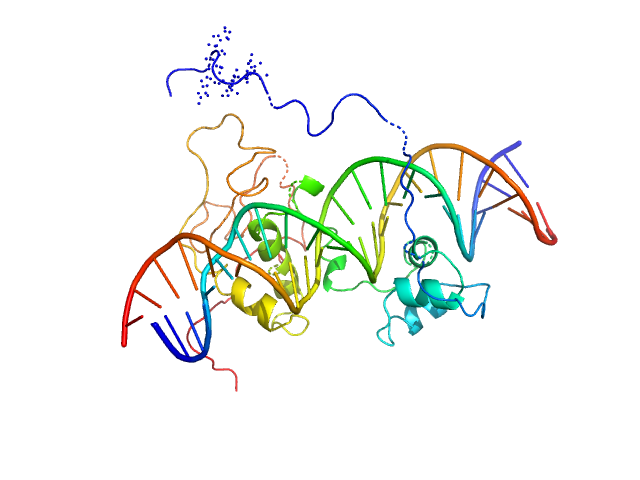
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
24mer dsDNA monomer, 15 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
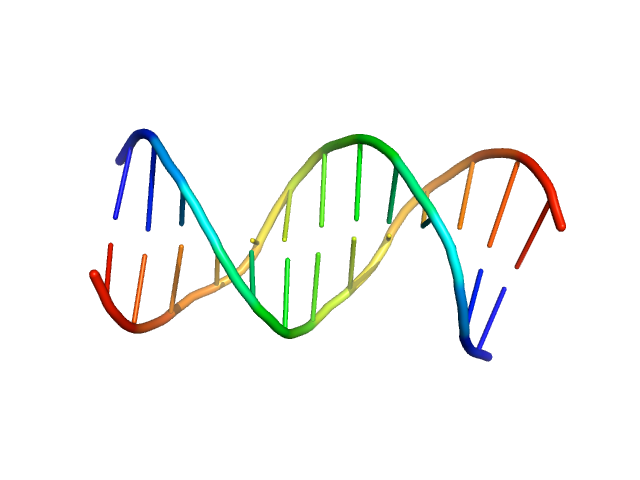
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
13mer dsDNA monomer, 8 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
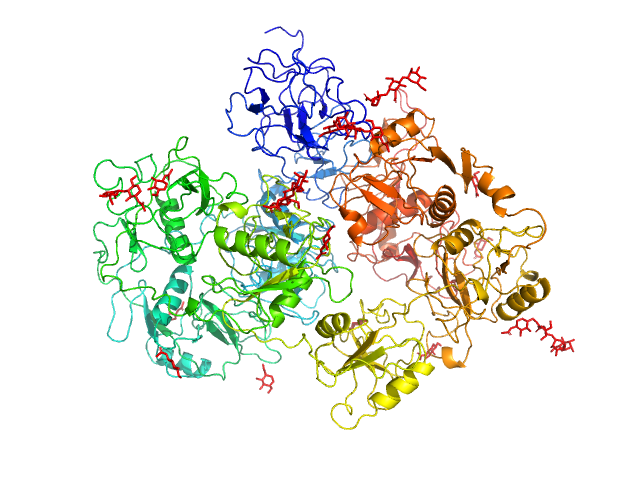
|
| Sample: |
Secretory phospholipase A2 receptor monomer, 189 kDa Homo sapiens protein
|
| Buffer: |
10 mM Bis-Tris, 150 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 19
|
Structure of PLA2R reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch
Proceedings of the National Academy of Sciences 119(29) (2022)
Fresquet M, Lockhart-Cairns M, Rhoden S, Jowitt T, Briggs D, Baldock C, Brenchley P, Lennon R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
426 |
nm3 |
|
|
|
|
|
|
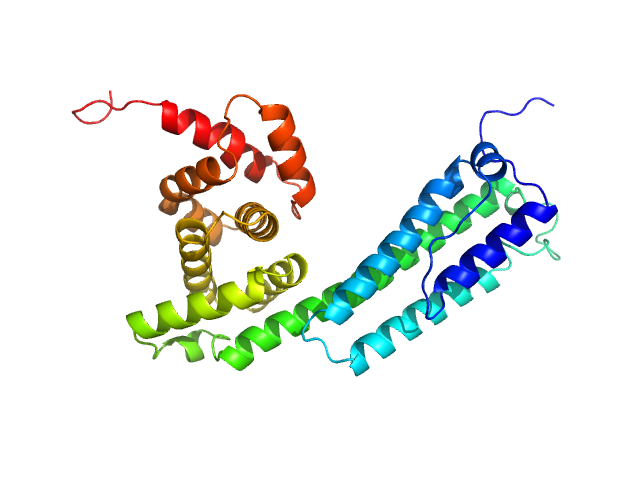
|
| Sample: |
Similar to Aedes aegypti 34 kDa salivary secreted protein 34k-2 monomer, 35 kDa Aedes albopictus protein
|
| Buffer: |
25 mM HEPES, 0.1 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Sep 3
|
A salivary factor binds a cuticular protein and modulates biting by inducing morphological changes in the mosquito labrum.
Curr Biol (2022)
Arnoldi I, Mancini G, Fumagalli M, Gastaldi D, D'Andrea L, Bandi C, Di Venere M, Iadarola P, Forneris F, Gabrieli P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
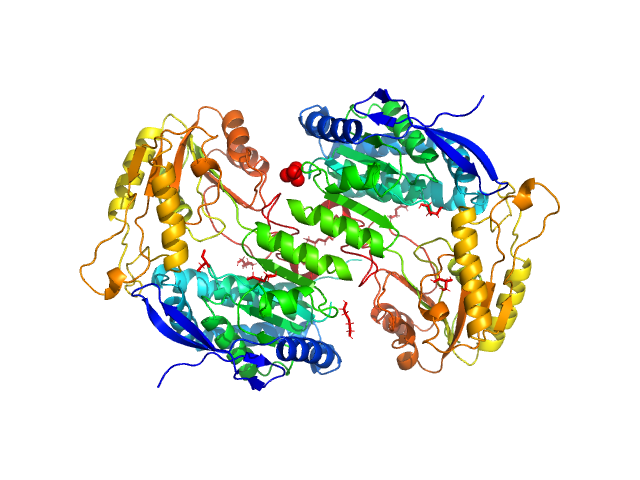
|
| Sample: |
2-aminomuconic 6-semialdehyde dehydrogenase dimer, 101 kDa Pseudomonas sp. protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Feb 1
|
The tetrameric assembly of 2-aminomuconic 6-semialdehyde dehydrogenase is a functional requirement of cofactor NAD(+) binding.
Environ Microbiol 24(7):2994-3012 (2022)
Shi Q, Chen Y, Li X, Dong H, Chen C, Zhong Z, Yang C, Liu G, Su D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
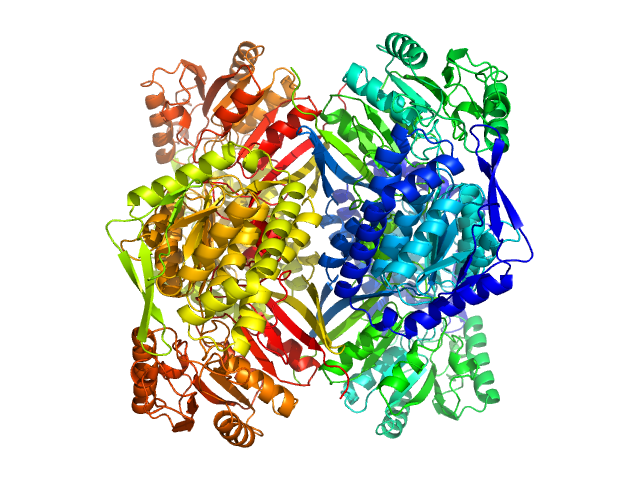
|
| Sample: |
2-aminomuconic 6-semialdehyde dehydrogenase tetramer, 215 kDa Pseudomonas sp. protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Feb 1
|
The tetrameric assembly of 2-aminomuconic 6-semialdehyde dehydrogenase is a functional requirement of cofactor NAD(+) binding.
Environ Microbiol 24(7):2994-3012 (2022)
Shi Q, Chen Y, Li X, Dong H, Chen C, Zhong Z, Yang C, Liu G, Su D
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
326 |
nm3 |
|
|
|
|
|
|
|
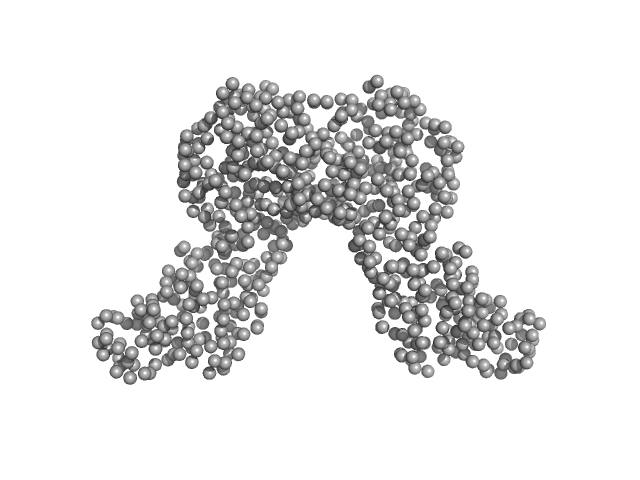
| Sample: |
Riboflavin biosynthesis protein RibD dimer, 80 kDa Acinetobacter baumannii protein
|
| Buffer: |
150 mM NaCl, 10 mM Tris, 1 mM DTT, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aconitate hydratase B monomer, 96 kDa Acinetobacter baumannii protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 1 mM DTT, 3% v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
150 |
nm3 |
|
|